Microbial Ecology Methods in Streams: From Cultivation to DNA Sequencing
Historical Limitations: The Great Plate Count Anomaly. Historically, studies of microbial diversity relied exclusively on cultivation-based methods, which severely underestimate true abundance and diversity. This long-recognized discrepancy is known as "The Great Plate Count Anomaly", a phenomenon impacting all microbial ecosystems but particularly problematic in understudied systems like streams. Consequently, decades-old studies based on cultivation failed to reveal the vast majority of the microbial community, as less than 1% of bacteria may be cultivatable, as reviewed by Nichols [6]. While cultivation provides valuable living stocks for physiological study, it does not accurately represent in-situ community composition or true microbial diversity, creating a significant gap in our historical understanding.
The Molecular Revolution and Its Challenges. The advent of high-throughput sequencing has fundamentally transformed microbial ecology, though some ecosystems like streams remain relatively understudied. The dynamic nature of streams, with their large temporal and spatial variations, presents challenges for developing broad generalities. A critical limitation of modern DNA-based methods is that they reveal the genetic potential for a function rather than the function itself. Specifically, detecting DNA from a particular microbial taxon in an environmental DNA sample does not confirm that the organism is viable, active, or expressing the gene of interest, decoupling community composition from immediate ecosystem function.
Microbial Groups and Their Habitats in Streams. Stream microbial communities are highly diverse, encompassing algae (especially diatoms and green algae), fungi (notably aquatic hyphomycetes), archaea, and bacteria. Due to their critical roles in processes like denitrification and decomposition via extracellular enzymes, bacterial ecology is widely studied from a functional standpoint, though less so from a taxonomic perspective. Research often focuses on specific stream habitats, primarily the water column and the benthos, including biofilms on surfaces like leaves, rocks, and wood. The character of a stream, influenced by location, climate, and substrata, directly impacts its microbial ecology; for instance, seasonal changes often affect planktonic bacteria more than benthic biofilm communities.
The Evolution of Methodological Approaches. Methodological approaches for assessing microbial community composition have evolved significantly over time, as summarized in Figure 1. Due to cultivation limitations, researchers now rely heavily on molecular methods. Initially, the field utilized DNA fingerprinting techniques like Terminal Restriction Fragment Length Polymorphism (T-RFLP) and Denaturing Gradient Gel Electrophoresis (DGGE). These PCR-based methods provide rapid measures of alpha-diversity (within a sample) and beta-diversity (between samples) by visualizing variations in specific genes. However, they describe communities in terms of Operational Taxonomic Units (OTUs) without revealing the taxonomic identity of the organisms, limiting the biological insights that can be drawn.
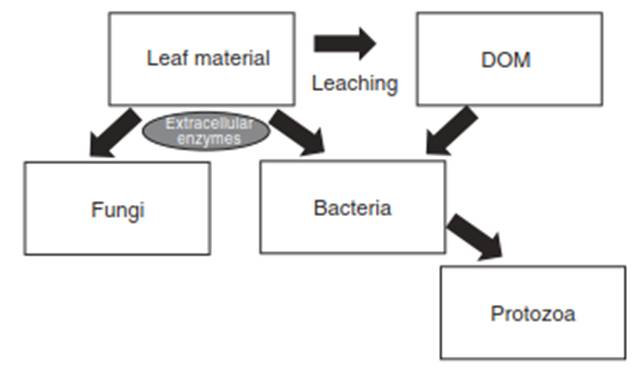
Figure 1. Basic changes in the methods used for the analysis of environmental DNA extracts over time
Limitations of PCR-Based Methods. Significant concerns exist with any PCR-based approach due to inherent PCR bias, where steps in the amplification process fail to provide an accurate quantitative representation of the microbial community. For example, primers may preferentially amplify one microbial group over another. This bias is most critical for groups that are underrepresented in sequence databases. Furthermore, when using a common target like the 16S rRNA gene for prokaryotes, the taxonomic resolution is limited, meaning that biologically relevant differences can exist between cells with identical sequences. These methodological constraints underscore the challenge of obtaining a truly unbiased census of microbial life.
The High-Throughput Sequencing Paradigm. The limitations of early methods were overcome by the invention of high-throughput sequencing technologies, such as pyrosequencing and the more recent Illumina sequencing[10]. This paradigm shift allows for the generation of massive amounts of sequence data in a rapid and cost-effective manner. Instead of just producing fingerprint patterns, sequencing provides the detailed information needed to name taxa by comparing sequences to existing databases. This technological leap moved the field's primary challenge from data generation to data processing, analysis, and interpretation, enabling more powerful genomic-level inquiries.
Expanding Targets: From Taxonomy to Function. Molecular methods can be characterized by their genetic target. Some target genes useful for taxonomy, such as the bacterial 16S rRNA gene, the fungal Internal Transcribed Spacer (ITS), or the 18S rRNA gene for diatoms and protozoa. Others target functional genes, like nosZ, which encodes for nitrous oxide reductase in the denitrification pathway. Crucially, methods can also target transcribed RNA (metatranscriptomics) to understand gene expression, overcoming the limitation of DNA-based methods that only indicate potential. Metagenomics and metatranscriptomics provide a comprehensive picture of the microbiome's potential and activity, even allowing for the discovery of novel genes.
Bioinformatics and Data Analysis Challenges. The generation of vast sequence data necessitates sophisticated tools for bioinformatics and statistical analysis. Two widely used, open-source bioinformatics pipelines are QIIME[12] and MOTHUR[13]. There is considerable debate within the scientific community regarding the best tool for different settings, with differences ranging from practical concerns to their impact on reported outcomes. As reviewed by Nilakanta et al. [14], these tools have distinct features and attributes. A critical question is whether different analytical approaches lead to the same biological conclusions; further study is needed, though research by Westcott and Schloss [15] suggests that analysis methods can affect OTU quality and stability. Consistency in analysis and data availability is crucial for facilitating meta-analyses to identify general ecological trends.
Date added: 2025-11-17; views: 115;
