The gene expression and repression
The genetic mechanisms of gene expression were studied by French scientists F. Jacob and J. Monod in 1961. The main statement of their discovery is that the genes may be of two kinds. One is structural genes, which encode information about macromolecules made by cell. Second is regulatory genes (or acceptory genes), which don’t encode polypeptide chain, but they regulate working of structural genes with help of different proteins attached to them.
Operon - is a cluster of functionally related genes transcribed onto a single mRNA molecule in bacterial cell. It is consist of structural genes and regulatory genes related to them. It represents a regulatory unit of gene expression. The structure and functioning of operons were studied on example of lac-operon of E.coli.
This operon is responsible for synthesis of protein that bacteria use to obtain energy from the sugar lactose. Operon is started from CAP site. It is a site for CAP-protein binding. CAP is an activator protein, which facilitates the unwinding of DNA duplex and so enables the polymerase to bind the nearby promoter. CAP protein need to be activated by existed in a cell cAMP, Next to CAP site is promoter. Promoter is nucleotide sequence recognizable by RNA polymerase. RNA polymerase binds promoter and then moves along operon transcribing it.
Next to promoter is operator. It is consist of 21 nucleotide pairs. It is a place for regulatory protein binding, which may suppress transcription. Next is a group of structural proteins. Operon is terminated by terminator. It is a short DNA region, which works as a stop signal for transcription (pic 6.2).
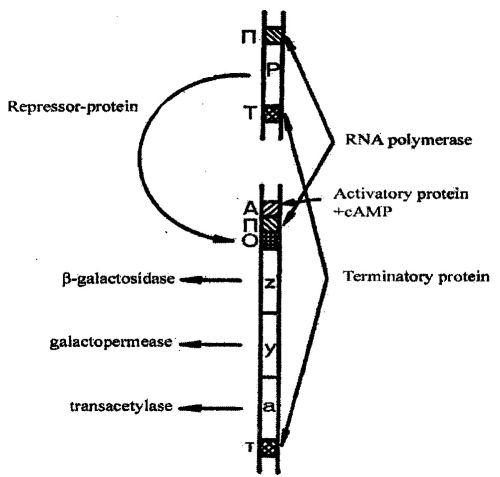
Pic. 6.2. The structure of lactose operon of E.coli: П - promoter; P - regulating gene; A - region for activating protein attachment; О - operator, T - terminator, z, y, a, - structural genes (by S. M. Gershenzon, 1979)
The main regulation of lac- operon working is performed by regulatory protein, which is encoded by regulator gene (pic 6.3). This protein continuously persists in a cell in very small amount. There are no more than 10 molecules of such protein in cytoplasm at the same time.
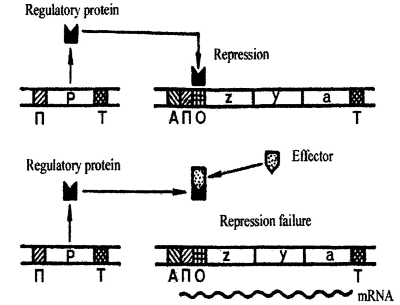
Ріс. 6.3. The repression (upper scheme) and induction (lower scheme) of lac-operone of E.coli (by M. S. Gershenzon, 1979)
This protein may bind the operator site of operon. Binding the repressor protein to the operator prevents binding of the polymerase to the promoter and so blocks the transcription of the structural genes of the operon. The synthesis of encoded enzymes fails. During lactose incoming, the repressor protein binds lactose and it changes its structure.
The repressor protein fails to bind operator site. Here, lactose works as an effector - a small substance that changes protein properties while binding with it. When operator is liberated from repression, RNA polymerase may moves along gene transcribing it. This starts a producing of all enzymes needed to lactose proceeding. That means gene induction. Thus, the regulation of lac-operon is performed by repressive protein binding to operator, which represses transcription. Induction occurs only when operator is free from repressive protein. This regulation type is called negative protein synthesis induction.
A negative repression has a similar mechanism. Negative repression is a regulatory protein binding to operator, which suppresses transcription. The differences in these two types are in following. During negative induction, an effector breaks regulatory protein binding to operator, whereas during negative repression effector enables regulatory protein to bind operator. An example of negative repression is the working of E.coli operon, which is responsible for tryptophan synthesis.
The regulator gene, which is not a part of tryptophan operon, always makes a regulatory protein. If cell use all tryptophan for its needs, regulatory protein doesn’t bind operator site. But if it is an excess of tryptophan in a cell, tryptophan binds regulatory protein modifying it structure. Modified protein enables to bind operator and suppress transcription of structural genes. Thus the tryptophan synthesis is terminated.
There is also a positive regulation of protein synthesis. The regulatory gene product activates operon transcription instead of repressing it. We may see this way of gene regulation in catabolic E.coli operon, which is responsible for producing enzymes for the arabinose sugar usage. The regulator gene produces regulatory protein, which binds with operon and activates its transcription.
During positive repression, the regulatory protein, encoded by regulator gene and activating operon transcription, fully or partially is suppressed by effector. This scheme may be useful for explaining tryptophan synthesis operon working in E.coli.
The table 6.1 summarizes main features of different type’s genetic regulation of operon.
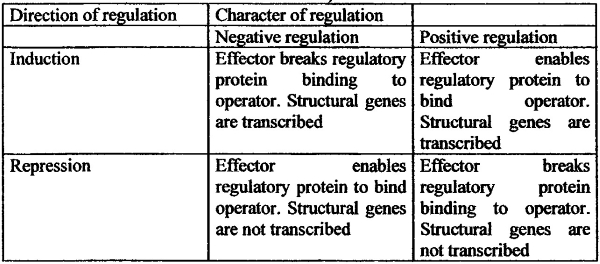
The table 6.1. The operon working regulation types, (by S. M. Gershinzon 1979)
The eukaryotes gene regulation has been studied less than prokaryotes gene regulation. It is due to complex gene structure, gene placing in chromosomes, nucleus having and cell differentiation. But it is possible that mechanisms of eukaryote gene regulation are similar to prokaryote one. But they have significant differences. Firstly, almost all eukaryotic genes contain only one structural gene, instead of several structural genes in bacterial operon. Secondary, in eukaryotes, the genes, which are responsible for different steps of one biochemical pathway, are spread throughout genome. Bacteria generally have them localized in one operon.
Thirdly, eukaryotes have a group simultaneous genes repression in whole nucleus, in whole chromosome or in significant region of it. It is mostly performed by histon proteins, which are a structural component of chromosomes. An example of it is full total repression of gene activity during spermatogenesis. Fourthly, the gene expression of eukaryotes may be regulated by steroid hormones (pic. 6.4).
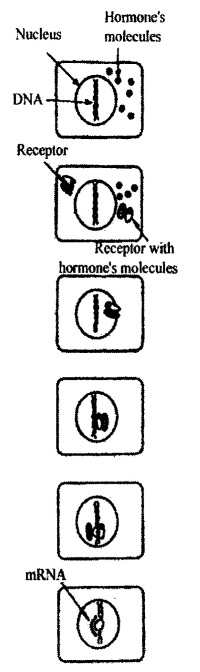
Ріс. 6. 4. The regulation of transcription by steroid hormones (by S. M. Oershenzon. 19791)
The target cells have special receptor proteins. These receptors are encoded by testicular feminization gene of X-chromosome. Binding of the receptor to hormone leads to complex formation. This complex activates expression of definite gene. Fifthly, eukaryotes’ genes may change their activity during ontogenesis.
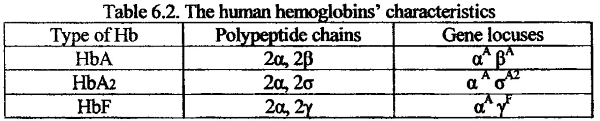
The example of different gene expression in ontogenesis is a genetic control of human hemoglobin synthesis. It is known that hemoglobin molecule contains four polypeptide chains: two identical α-chains and two identical ß-chains. The adult hemoglobin (HbA) is differed from embryo hemoglobin (HbF). The differences are related withß-chains. In embryo’s hemoglobin there is noß-chain. It is replaced byY-chain. However, in an adult blood we may find an HbA2 in small amount.
The ß -chain in this hemoglobin is replaced by ơ-chain. All three types of normal human hemoglobins are encoded by separate locuses. The locusα A is responsible for a-chain synthesis. It is active throughout all life. The locusß A is responsible for polypeptide chains synthesis in HbA. It becomes activated only after birth. The locuY F is responsible for polypeptide chains synthesis in HbF. It works actively during embryonic development. The locusy A2 is responsible for polypeptide chains synthesis in HbA2. It is active throughout the life after birth.
Each of hemoglobin genes (a Aß A,Y F,y A2) is a structural gene because it is responsible for primary structure of polypeptide chain.
We see different kinds of hemoglobins that arise from different gene combinations. It is clear that working of structural genes is under supervision of regulatory genes. It is become evident from a fact of HbF exchange to HbA after birth. Here we see the working of special gene - “switch gene”, which suppress activity ofγF gene and activateß A gene. As result of this, embryo hemoglobin is exchanged to adult one. We may suppose that this simultaneous switching of gene activity may be due to action of gene operator of bothy F and ßA genes.
Date added: 2023-01-09; views: 612;
