Nucleic Acid Structure
After Avery's work, nucleic acids came under prompt and intense study. They were quickly found to be large molecules. The illusion that they were small came about because earlier methods of extraction had been harsh enough to break up the molecules into smaller fragments as they were being extracted. Gentler techniques extracted nucleic acid molecules as large as or larger than the largest protein molecules.
The Austrian-American biochemist, Erwin Chargaff (1905- ), broke down nucleic acid molecules and subjected the fragments to separation by paper chromatography. He showed, in the late 1940s, that in the DNA molecule, the number of purine groups was equal to the number of pyrimidine groups. More specifically, the number of adenine groups (a purine) was usually equal to the number of thymine groups (a pyrimidine), while the number of guanine groups (a purine) was equal to the number of cytosine groups (a pyrimidine). This might be expressed as A = T and G = C.
The New Zealand-born British physicist, Maurice Hugh Frederick Wilkins (1916- ), applied the technique of X-ray diffraction to DNA in the early 1950s, and his colleagues at Cambridge University, the English biochemist, Francis Harry Compton Crick (1916- ) and the American biochemist, James Dewey Watson (1928- ), attempted to devise a molecular structure that would account for the data obtained by Wilkins.
Pauling had just evolved his theory of the helical structure of proteins, and it seemed to Crick and Watson that a helical DNA molecule would fit in with Wilkins' data. They needed a double helix, however, to account for Chargaff's findings as well. They visualized the DNA molecule as consisting of two sugar-phosphate backbones winding up about a common axis and forming a cylindrical molecule. The purines and pyrimidines extended inward from the backbones, approaching the center of the cylinder. To keep the diameter of the cylinder uniform, a large purine must always be adjacent to a small pyrimidine. Specifically, an A must adjoins a T and a G must adjoin a C and it is thus that Chargaff's findings were explained.
Furthermore, an explanation was now available for the key step in mitosis, the doubling of the chromosom.es (and for a related problem as well, the manner in which virus molecules reproduced themselves within a cell). Each DNA molecule formed a replica of itself ("replication") as follows: The two sugar-phosphate backbones unwound and each served as a model for a new "complement." Wherever an adenine existed on one backbone, a thymine molecule was selected from among the supply always present in the cell, and vice versa; wherever a guanine molecule was present, a cytosine molecule was selected, and vice versa. Thus, backbone i built up a new backbone 2, while backbone 2 built up a new backbone 1. Pretty soon, two double helices existed where only one had before.
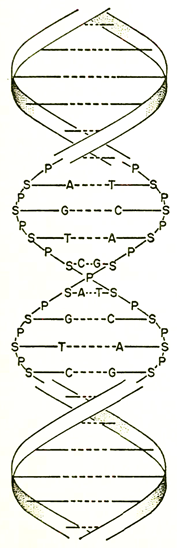
The double helix of the DNA molecule. The backbone strands are made up of alternating sugar (S) and phosphate (P) groups. Extending inward are the bases, adenine (A), guanine (G), thymine (T), and cytosine (C). The dashed lines are hydrogen bonds which link the strands. In replication, each of these strands will produce its complement from the purines and pyrimidines (A, G, C,T, etc.) that are always present in the cell. (After a drawing in Scientific American.)
If DNA molecules did this all along the line of a chromosome (or virus), one ended with two identical chromosomes (or viruses) where only one had existed before. The process was not always carried through perfectly. When an imperfection occurred in the replication process, the new DNA molecule was slightly different from its "ancestor"; and one had a mutation.
This Watson-Crick "model" was announced to the world in 1953.
Date added: 2022-12-11; views: 954;
