Pseudomonas Diversity, Genetics, and the Distinct Features of Pseudomonas mendocina
The genus Pseudomonas comprises an exceptionally diverse group of bacteria, inhabiting a vast range of terrestrial and aquatic environments. While some members are serious pathogens of animals and plants, others are free-living saprophytes crucial to nutrient cycling (Figure 1) [1]. This remarkable physiological diversity is underpinned by a famously high degree of genetic plasticity, facilitated by horizontal gene transfer. This adaptability allows pseudomonads to acquire genes that confer survival advantages in new and challenging environments, leading to the evolution of numerous strains with specialized metabolic capabilities. Consequently, many Pseudomonas species have become invaluable tools in biotechnology and bioremediation, and their genetic tractability makes them highly amenable to laboratory modification for further customization.
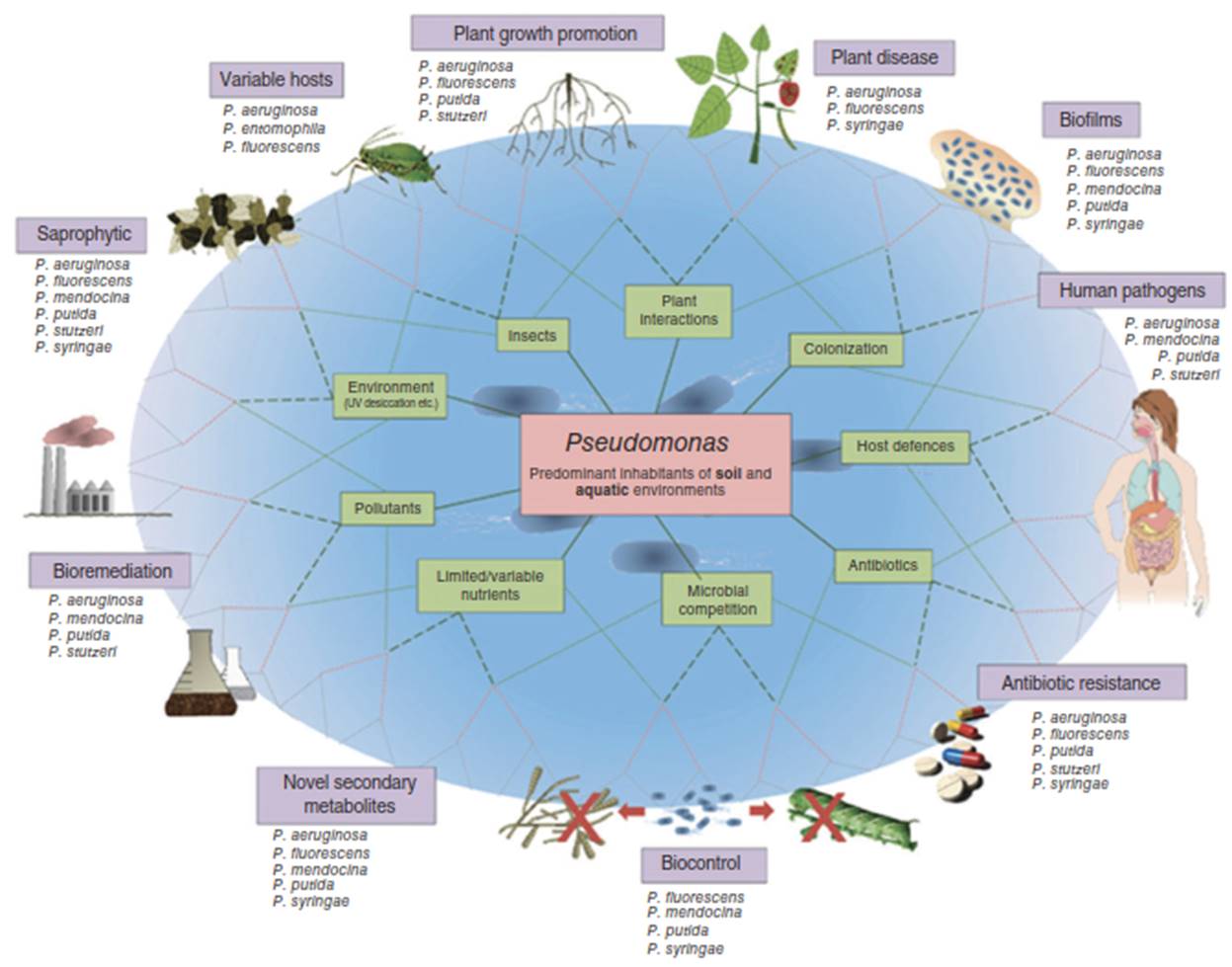
Figure 1. Metabolic and ecological diversityof Pseduomonas spp. Source: Used with permission from Ref
What Is a Pseudomonad? The genus Pseudomonas was first proposed in 1894, with mid-twentieth-century descriptions being intentionally broad to accommodate a wide range of known bacteria. A typical pseudomonad was historically described as a motile, rod-shaped, Gram-negative bacterium that employs respiratory metabolism, typically using oxygen or alternatively nitrate as a terminal electron acceptor. This loose definition made the genus a catch-all for many motile, non-fermenting Gram-negative rods. The advent of molecular taxonomy in the late 20th century, however, led to significant reorganization [4-6]. A landmark 1973 study by Norberto Palleroni utilized ribosomal RNA (rRNA) sequencing to demonstrate that the genus comprised five distinct groups [7].
Only Group I contained the true core of the genus, including the type species Pseudomonas aeruginosa and its close relatives. This group encompasses both fluorescent species like Pseudomonas fluorescens, Pseudomonas putida, and Pseudomonas syringae, and non-fluorescent species such as Pseudomonas stutzeri and Pseudomonas mendocina. The species in the other rRNA groups (II-V) were subsequently reclassified into separate genera within the Proteobacterial phylum [8]. Despite this taxonomic refinement, genomic variation within the true Pseudomonas genus remains immense. Different species, and even different strains of the same species, often possess highly variable genomes, a key feature of their adaptability [9] (Figure 2).
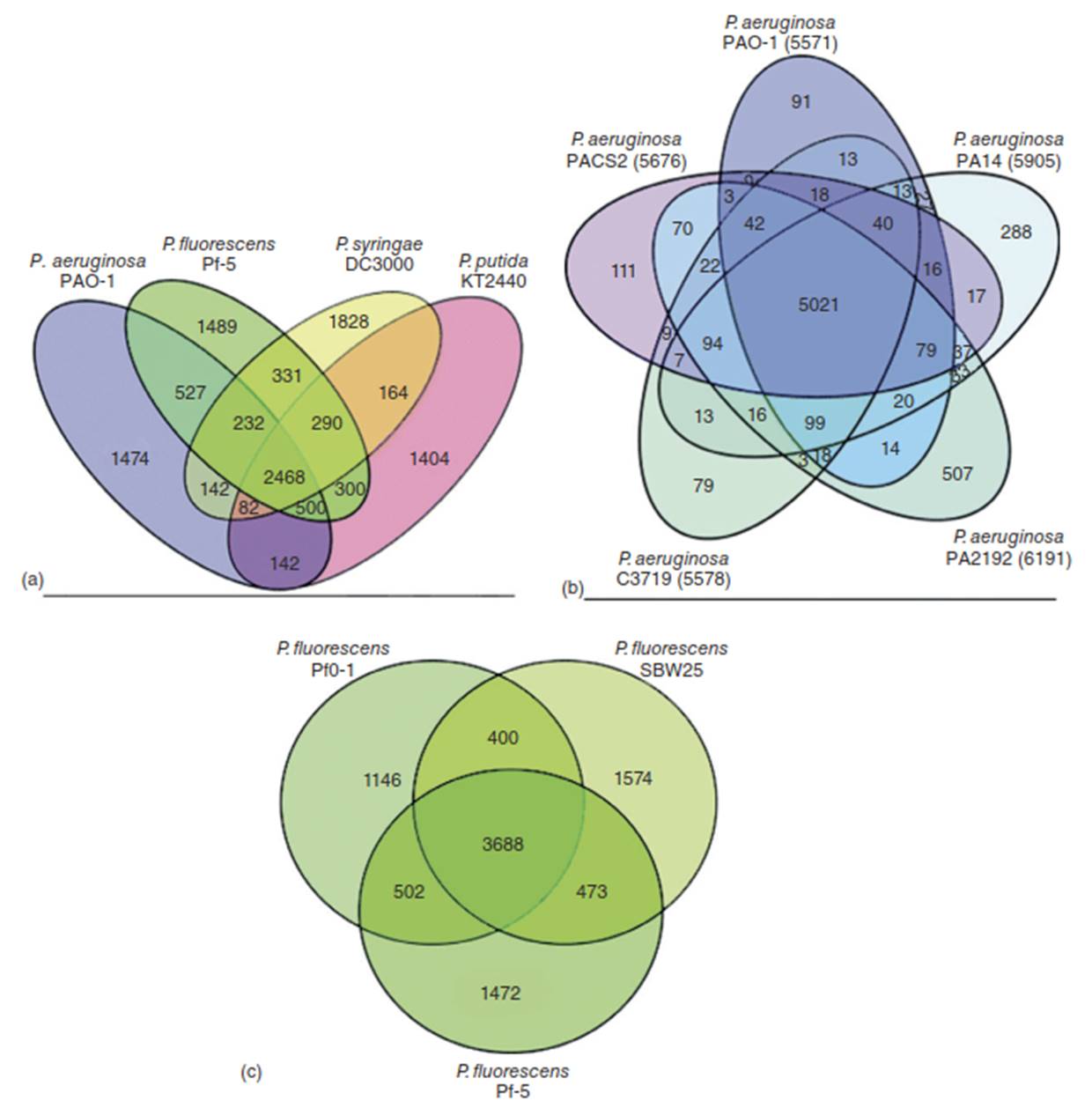
Figure 2. Pseudomonas spp. exhibit remarkable metabolic diversity. As an example, these Venn diagrams showthe number of proteins shared or unique among (a) four Pseudomonas species, (b) five strains of P aeruginosa, and (c) three strains of P.fluorescens. Source: Reproduced from Ref. [9] with permission from The Royal Society of Chemistry
An analysis of the growing number of sequenced Pseudomonas genomes reveals significant diversity. Genome sizes range from 3.1 to over 7 megabases, encoding between approximately 4,000 to more than 6,000 genes. The estimated size of the core genome—the set of genes shared by all members—shrinks as more genomes are compared. For instance, a comparison of four key strains (P. aeruginosa PAO1, P. fluorescens Pf-5, P. syringae DC3000, and P. putida KT2440) showed less than half of their genes were common, and a study of 14 genomes reduced the core set to around 1,700 genes [1, 10]. This genomic diversity is reflected in the vast array of secondary metabolites produced, including antibiotics, siderophores, surfactants, and virulence factors, which are essential for niche adaptation and valuable for biotechnological applications.
Distinguishing Features of Pseudomonas mendocina. The species Pseudomonas mendocina was discovered serendipitously during efforts to characterize P. stutzeri, when researchers noted that some isolates could not utilize starch or maltose as carbon sources [11]. These unique isolates, sourced from terrestrial and freshwater environments in Mendoza, Argentina, were subjected to further genomic and rRNA analysis. The results confirmed they were sufficiently distinct from P. stutzeri to warrant designation as a new species, leading to the birth of P. mendocina, with the Palleroni strain as its first named member.
Morphologically and metabolically, P. mendocina shares several traits with its relative P. stutzeri. It is a Gram-negative, rod-shaped, non-fluorescent, metabolically respiratory γ-Proteobacterium. It primarily uses oxygen as an electron acceptor, with nitrate (NO3- ) as a secondary option, and its optimal growth temperature is near 30°C. However, its inability to metabolize maltose or starch was the initial distinguishing characteristic. Furthermore, its colonial morphology is distinct; while P. stutzeri forms bumpy, pale colonies, P. mendocina colonies are typically flat, smooth, and yellowish due to the production of carotenoid pigments.
Strains of P. mendocina have since been isolated from diverse locations worldwide, indicating a widespread distribution [3]. To date, eight strains have been submitted for whole-genome sequencing, revealing a median genome size of 5.29 megabases, encoding about 4,659 genes with a high GC content of 62.75%. Unlike other well-studied Pseudomonas species, detailed comparative genomic analyses of P. mendocina strains are not yet available. The submitted strains reflect research interests in various biotechnological applications, which can be categorized into three main areas: metal resistance and mobilization (e.g., strains S5.2, S13.2, ymp); reductive detoxification of nitrogen oxides (strain DLHK); and the metabolism of diverse carbon sources (strains ZWU0006, NK-01, EGD-AQ5). Although clinical infections are exceedingly rare [19], its ability to live commensally within some animals demonstrates its ecological versatility [17].
Date added: 2025-11-17; views: 124;
