DNA: The blueprints of living cells
A blueprint is used for storing information about the construction and maintenance of a factory and the processes inside.
Blueprints for the building may have information about the structural components (e.g. brick outer shell supported by iron beams), the ventilation systems (e.g. made of aluminum ducting), the complex electrical configurations (e.g. different voltages, wires, and location of the outlets). Blueprints can also determine how a building is built. Is a crane needed? If so, scaffolding may be erected to accommodate for this.
Blueprints can also be used to define the equipment inside, how it is positioned, and the speed at which it operates. A lot of equipment and infrastructure is permanently connected to the factory. In this case, the components, machines and the factory building itself meld to become one. In other words, a factory is not just a building, but is the building, equipment, and even the people inside, turning raw materials into end products.
Why are we talking about factories? Factories as we know them are analogous to living cells—except they are much larger and made of different materials. The blueprints that define how to build and maintain a factory are comparable to the blueprints of living cells. In the case of living cells, however, there is not a miniature piece of blue paper rolled up in every cell. There is a microscopic chemical string, the DNA.
Unlike a factory blueprint that you can see with your eyes, hold in your hands, and is written in a language that is well understood, the blueprint of a living cell, DNA, is a microscopic “chemical string” of nucleotide building blocks. This string has a distinct “language” that all living cells know how to read and write. Only since 1953 when Francis Crick, James Watson, Rosalind Franklin, and Maurice Wilkins discovered the structure of DNA have we begun to understand it and its language. Here is some of what we know:
DNA is made of four chemical building blocks called nucleotides that are attached to one another in various orders to form a “string” of the nucleotides that can be thousands to millions of nucleotides long. Each nucleotide is made of CHOPN, and each nucleotide is made up of three “sub-molecules” called a phosphate, a nitrogenous base, and a deoxyribose sugar (Figure 1-16). Have a look at Figure 1-16, do you see the CHOPN chemical elements?
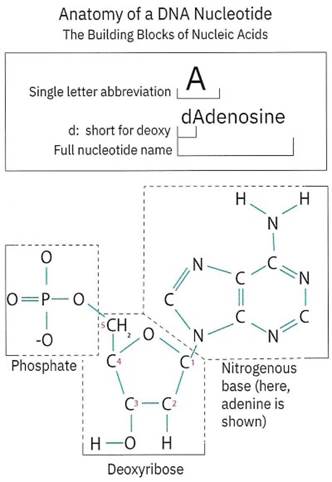
Figure 1-16. The building blocks of DNA, nucleotides, have important characteristics. A conserved phosphate, deoxyribose sugar, and the variable nitrogenous base
Phosphate is a molecule with a phosphorus atom bound to four oxygen atoms. Phosphate is a critical component of the “sugar-phosphate” backbone of DNA (Figure 1-17). The phosphate group is very negatively charged and is what gives DNA an overall negative charge. As you saw in Chapter l’s exercise, the negative charge of DNA is an important chemical characteristic that you took advantage of to extract DNA from cells using chemistry.
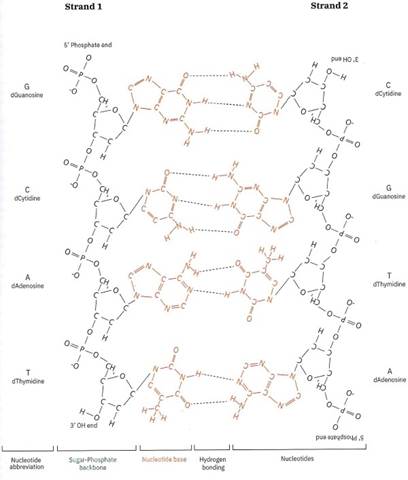
Figure 1-17. Nucleotides are strung together into strands via the sugar-phosphate backbone, Two strands of DNA can zip together to form a double helix. They are able to zip together because of a special bonding force called hydrogen bonding. The hydrogen bonding occurs between the nitrogenous bases alone, not via the sugar-phosphate backbone
Nitrogenous bases are the variable part of a nucleotide. While every nucleotide has the same phosphate backbone, there are four different nitrogenous bases in DNA - Guanine, Thymine, Adenine, and Cytosine (Figure 1-18). Can you spot the different nitrogenous bases? If you have trouble, try the What is DNA? application mentioned in the hands-on exercise. The nitrogenous base is the part of the nucleotide molecules that is the information that the cell machinery “reads”.
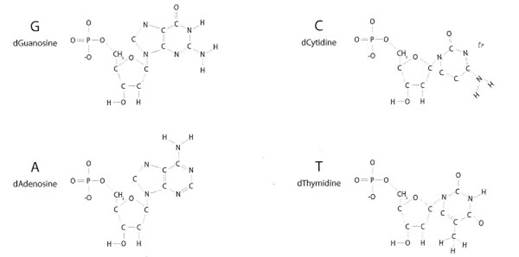
Figure 1-18. The four nucleotide building blocks of DNA are called deoxyguanosine (dG), deoxycytidine (dC), deoxyadenosine (dA), and deoxythymidine (dT). For simplicity, they are often referred to by their nitrogenous base names: guanine (G), cytosine (C), adenine (A), and thymine (T). A small d is an abbreviation for ‘deoxy.’
Deoxyribose is a sugar ring that is the “D” in DNA. Deoxyribose connects to both the nitrogenous base and the phosphate (Figure 1-16). When multiple nucleotides are connected, as seen in Figure 1-16, you’ll see that the deoxyribose also connects to the phosphate of the next nucleotide. The deoxyribose and the phosphate together form the “sugar-phosphate” backbone of DNA (Figure 1-17).
Nucleotides are usually referred to by the first letters of their nitrogenous base names. Because four different nitrogenous bases can be attached to the deoxyribose ring, four different possible nucleotides make up DNA: A for adenine, T for thymine, C for cytosine and G for guanine (Figure 1-18). You can see two nucleotide chains side-by-side in Figure 1-17, and both have each of the four nucleotides.
During regular cell activity, millions of each of the four nucleotides are produced by the cell. The nucleotides in the cell can be collected by a cell machine called DNA polymerase, which strings them together like pearls in a necklace. It is the order of the nucleotides ...ATGGCGGTTACC... which we call the DNA sequence that the cell machinery reads and understands.
As you see in Figure 1-17, the sugar-phosphate backbones are on the outside of the molecule, and the nitrogenous bases are buried inside. If you imagine DNA to be a ladder, the sugar-phosphate would be the outside of the ladder, and the nitrogenous bases would be the ladder rungs. This means that the “information” that the cell’s machinery reads is in the rungs of the ladder. If you look at Strand 1 in Figure 1-17, reading from the top “5’ Phosphate end” down to the “3’-OH end”, the DNA sequence is GCAT.
When two strands of DNA come together, they form a three-dimensional structure known as the double helix. Looking down at the structure from the top, it looks like a spiral staircase (Figure 1-19). The blue- gray region in the center of the illustration shows the nitrogenous bases, the A’s, T’s, C’s and G’s that form the information layer of the DNA blueprint.
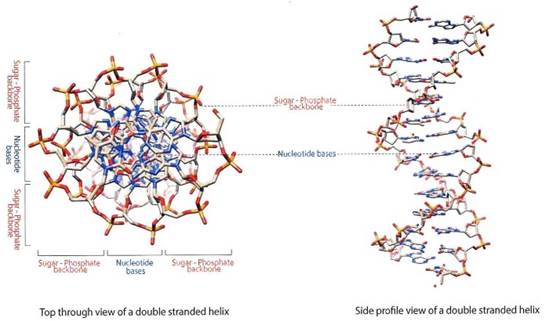
Figure 1-19. The DNA helix is a three-dimensional structure that forms when two strands of DNA bind together using Chargaff’s Rule. The blue-gray region indicates the nitrogenous bases, and the red-orange-gray region indicates the negatively charged sugar-phosphate backbone
These are held in place by the outer orange-red-gray region of the negatively-charged sugar-phosphate backbone. The sugar-phosphate backbone has two critical functions: i) to maintain structure and hold the nucleotide bases in place; ii) to selectively attract cellular machinery called proteins so that the cell can “read” the DNA. In simplest terms, you can think of the sugar-phosphate backbone as the paper of a blueprint, and the nitrogenous bases as the words (information) written on the blueprint that can be understood.
A cool property of DNA is that the two strands of a double helix are only loosely connected by hydrogen bonds (which you will learn more about in Chapter 6). These loose bonds mean that the two strands can come apart like a zipper on a jacket. On a jacket, each strand of the zipper is quite strong, whereas the interaction between the two zipper strands can be undone easily with a simple pass of the zipping mechanism. In DNA, the chemical bonds in the sugar-phosphate backbone are very strong, but the bonds between two strands are relatively weak (Figure 1-17; dashed lines).
This “unzipping” capability is a key feature of how DNA functions. The cellular machinery can bind to the sugar-phosphate backbone and pull the double helix apart, unzip to get the information as needed, and when finished, the molecule closes up and protects the information.
Are there any rules on how two DNA strands can zip together? Yes! One fundamental rule was discovered by a famous scientist called Erwin Chargaff. Chargaff’s rule states that: Only an “A” in one strand can bind to a “T” in the other, and only a “C” in one strand can hind to a “G” in the other: the nitrogenous bases of A’s and T’s complement each other and the same with C’s and G’s. In Figure 1-17, you’ll see Chargaff’s Rule in action. Only when two strands have “complementary” nucleotides can they wind up together! As you learn more about the nuts and bolts of genetic engineering in later chapters, this will become much more evident.
Date added: 2023-11-02; views: 621;
