Sequencing through DNA Synthesis
The next step in the sequencing process is the physical separation and PCR amplification of individual DNA molecules in the DNA fragment library prepared above. This is done through the use of the ligated adapters. There are two main methods for physical separation and amplification. The first uses very small beads which are coated with the complement oligonucleotide to the ligated adapter sequence.
Beads and denatured library DNA are mixed in a defined ratio to ideally achieve a single DNA molecule hybridized to each bead. Excess DNA is washed away and the beads are added to an oil- aqueous emulsion in such a way as to have a single bead in each aqueous bubble.
Also contained in this mixture are a heat stable polymerase, dNTPs, and primers that hybridize to the library adapter sequence. Each aqueous bubble can be thought of as an individual PCR reaction chamber. During PCR, copies are made of the individual sequence.
These are contained within the aqueous bubble and re-hybridize to the target bead. After PCR is performed, each bead contains many identical copies of the individual DNA molecule that was originally hybridized to the bead. The second approach uses a single solid support coated with the complement to the ligated library DNA adapter.
Library DNA is denatured and hybridized to this solid support. An amplification reaction is then performed. When new DNA molecules are synthesized from their captured template they hybridize back onto the solid support adjacent to their parent. Through multiple iteration of amplification, tiny islands of DNA, all derived from the same sequence, are created.
The next step in this process is sequencing of the DNA bound to each individual bead or from each isolated DNA island (fig. 9). Pyrosequencing is ideally suited to the bead-based approaches (Harrington et al., 2013). The beads are loaded into a honeycomb-welled plate. Each well is loaded to contain an individual bead; each bead is coated with a unique clonal DNA sequence.
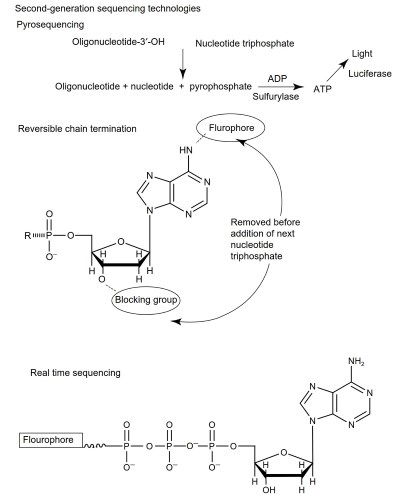
Figure 9. Methods for measuring DNA sequence analysis by synthesis
A deoxynucleotide (e.g., dATP), ADP, sulfurylase, luciferin, firefly luciferase, and a DNA polymerase are added to the plate. If the nucleotide (e.g., dATP) is incorporated by the DNA polymerase into the sequence attached to the bead, pyrophosphate is released. Pyrophosphate is converted to ATP by sulfurylase using ADP as a substrate.
Luciferase then takes the substrates luciferin and ATP to produce light. In short, if the deoxynucleotide is incorporated into the DNA attached to a bead, the well containing that bead will glow. From this, it can be inferred that the sequence starts with the complement to the nucleotide added (i.e., if dATP was added, the sequence starts with a T).
Next dCTP is added using the same approach, with the same result; a glowing well indicates that the next nucleotide in the target sequence is a G. Next comes dGTP and then dTTP. This cycle is repeated hundreds of times. Each time, a glowing well indicates the incorporation of the added nucleotide. Raw data are literally the amount of light produced by each well in each cycle.
From this, the sequence of each bead- attached clonal DNA population can be inferred. In a single plate, millions of individual beads can be sequenced, resulting in hundreds of millions of sequenced nucleotides from the short 100-200 bp fragment sequence library.
The analysis of millions of individual 'reactors' is why this approach is often referred to as 'massively parallel' DNA sequencing. Newer bead- based sequencing approaches can detect a change in pH in each reactor well that occurs with the incorporation of a nucleotide and associated release of pyrophosphate. This simplifies the chemistry behind the sequencing process.
For the solid support approach, modified dNTPs are used in the sequencing reaction. These modifications include a 3'OH blocking group and a fluorophore. The cycle of sequencing is conceptually very similar to the bead-based approach. To the solid support, a DNA polymerase and a single modified dNTP (e.g., dATP) is added. If the dATP is incorporated the tiny DNA island will fluoresce. A picture is taken and the blocking agent and fluorophore are chemically removed.
Each dNTP is added in turn (ddAPT, ddCTP, ddGTP, ddTTP) and the process is cycled hundreds of times. The end result is an inferred sequence based on the fluorescence, or lack thereof, in each cycle for each clonal DNA island. As with the bead-based approach, this can produce millions of individual 100-200 bp long sequence reads, one for each DNA island.
Date added: 2024-06-13; views: 466;
