Regulation of Cholesterol Biosynthesis
The complex regulation of cholesterol biosynthesis takes place at several levels. The rate of synthesis is highly responsive to cellular level of cholesterol. There exists a feedback regulation mediated by changes in HMGCR. High cholesterol levels cause reduction in transcription of HMGCR gene, leading to the reduction in the amount of HMGCR (messenger RNA) mRNA. HMGCR activity is also regulated through phosphorylation.
AMP (adenosine monophosphate)-dependent protein kinase (AMPK) catalyzes the phosphorylation and inhibition of the enzyme (Motoshima et al., 2006). Various cholesterol metabolites such as oxidized derivatives of cholesterol as well as cholesterol biosynthesis intermediates such as mevalonate and farnesol are additional components of the negative feedback loop that regulates the levels of HMGCR (Correll et al., 1994).
Cholesterol can also induce ubiquitination and degradation of HMGCR. Increases in cellular cholesterol concentrations triggers binding of HMGCR to the ER membrane proteins Insig-1 and Insig-2. This binding leads to the recruitment of gp78, the ubiquitin ligase that leads to ubiquitination of the enzyme. In this manner, HMGCR is marked for degradation and transported to proteasomes for degradation (DeBose-Boyd, 2008).
A complex of proteins that sense cellular cholesterol levels also regulates the rate of cholesterol synthesis. Sterol response element binding protein (SREBP), the SREBP cleavage activating protein (SCAP), and two SREBP-specific proteases (S1P and S2P) are the key components of this pathway (Horton et al., 2002; Bengoechea-Alonso and Ericsson, 2007). SREBP precursor proteins are embedded in the membrane of ER.
The N-terminal domain of SREBP acts as a transcription factor, whereas the C-terminal domain interacts with C-terminal domain of another ER protein SCAP, which contains a sterolsensing domain. As shown in Figure 4, when sterol levels are high, SCAP interacts with ER membrane protein, insulin regulated protein (Insig) (Yang et al., 2002).
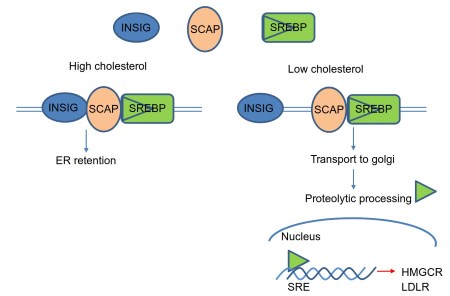
Figure 4. SREBP and SCAP in transcriptional control of cholesterol synthesis
Association with Insig leads to the retention of SREBP-SCAP complex in the ER (Yang et al., 2002). When sterol levels are low, SCAP and Insig do not interact, which leads to a conformational change in SCAP (Brown et al., 2002). The SREBP-SCAP complex then translocates from ER to Golgi apparatus (Figure 4).
Two proteases, S1P and S2P, act on SREBP in the Golgi apparatus (Nohturfft et al., 2000). SREBP is first cleaved by protease S1P, yielding a product that then acts as a substrate for protease S2P. This cleaved SREBP is then released to the cytosol and travels to the nucleus where it binds to sterol response element (SRE) DNA sites resulting in increased transcription of several genes involved in cholesterol synthesis pathway (Bengoechea-Alonso and Ericsson, 2007).
Date added: 2024-06-13; views: 493;
