Description of How the Components of the Lipidome Are Usually Analyzed
The large number of lipids and the high degree of structural variation among, and even within, the eight categories makes their analysis much more challenging than for most other categories of metabolites.
While it has been possible to profile some lipids by common structural features, such as glycolipids that can be captured by lectins that bind the carbohydrate moiety, for a more comprehensive lipidomic analysis it is necessary to use a method that is highly structure specific - and preferably can also be quantitative - such as mass spectrometry (MS).
The power of mass spectrometry is that compounds can be detected in very small amounts once they are ionized, and instruments are available to resolve ions by the mass to charge ratio (m/z), sometimes with high enough mass accuracy to match the data with the molecular formula of the corresponding lipid(s).
The analysis might not need such high mass accuracy if the compounds of interest can be distinguished otherwise (e.g., by their behavior on chromatography prior to mass spectrometry); however, the experimenter must always validate that the ions truly reflect the compound that it has been presumed to be.
This is even important when the instrument has a high mass accuracy (such as a time-of-flight, ToF, or fourier transform ion cyclotron resonance, FT-ICR mass spectrometers) because many lipids are isomeric (such as glucosylceramides vs galactosylceramides, which vary only in the stereochemistry of one of the hydroxyls on the carbohydrate) and/or are isobaric (i.e., have the same mass but different composition) within the accuracy of the analysis.
Additional information can be acquired about the compound from other features of the mass spectrum, such as the isotopic distribution, evidence that the compound is present as multiply charged species, etc. In metabolism studies, one can also introduce compounds that are enriched in stable isotopes and track these into downstream metabolites.
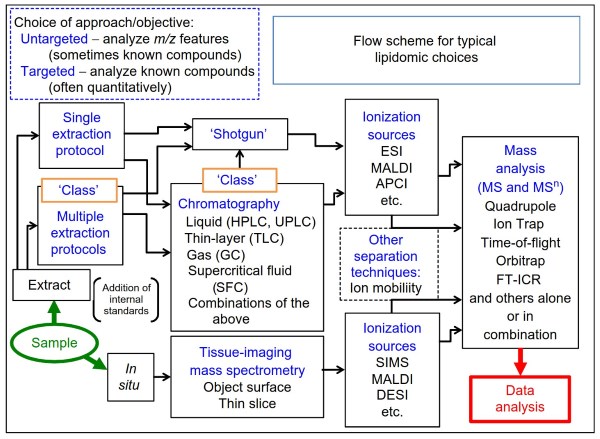
Figure 2. A flow diagram for the types of technical choices that can be used for a lipidomic analysis. Depending on the sample and the information desired, one might elect to extract the lipids or analyze them in situ. If extracted, the compounds can be analyzed using a ‘one-size- fits-all’ extraction protocol or with multiple extraction schemes that ensure high recoveries for all lipidomic categories (this has been called the CLASS approach for comprehensive lipidomics analysis by separation simplification).
The extracts can be analyzed by a ‘shotgun’ approach, which often involves introducing the entire sample into the mass spectrometer and having it conduct a broad, but high resolution, scan to identify as many compounds as possible, or if comparing two samples, features in the spectra that differ and might be interesting to pursue.
In the chromatography toolkit, a variety of methods can be used to partially separate the lipids to minimize ionization suppression of minor species as well as to separate compounds that cannot be distinguished by mass spectrometry alone (such as isomers). In the mass spectrometry toolkit, ion trap (which includes quadrupole and orbitrap), time-of-flight, and FT-ICR can be used alone or in combination with one another.
If analysis in situ is desired, this can be conducted by methods that ablate ions from the surface of the material directly (such as DESI and SIMS) or by first impregnating the sample with a matrix material that can be excited by a laser to generate ions (MALDI)
Lipidomic analysis requires a myriad of choices that must be made, and these steps will be discussed here and in the following sections of this article. The critical choices are summarized in Figure 2 and additional information can be found in excellent reviews (Murphy and Axelsen, 2011) and monographs on this topic (Brown, 2007). The first depend on the goals of the study: Is quantitation of the lipids necessary? If so, then a targeted LC-MS/MS protocol with multiple extractions and internal standards will probably be required.
Is the goal to discover potential biomarkers by surveying a large number of samples? If so, then an untargeted approach might be the best choice. Is it important to know the location of the analyte(s) in a sample such as a histological slice? If so, then an imaging MS method is usually selected.
Date added: 2024-06-13; views: 443;
