Signaling Lipids are Spatially Restricted and Scarce
Tight regulation of signaling lipids is reflected in their low abundance, even in tissues that specialize in processes that depend on bioactive lipids. For example, phospholipids represent approximately 65% of total cellular lipids, but the combined pool of phospholipid-derived signaling lipids may represent less than 1% of that value. However, quantitative methods such as mass spectroscopy have revealed general patterns in the relative abundance of many bioactive lipids.
For example, DAG and PtdOH are approximately an order of magnitude more abundant than the phosphatidylinositols, with PI(4,5)P2 and PI(4)P contributing approximately 61% and 37% or the PIPs, respectively (Guillou et al., 2007). Overall, it has become clear that the location and abundance of signaling lipids is carefully regulated to maintain coherent signaling networks (Figure 2).
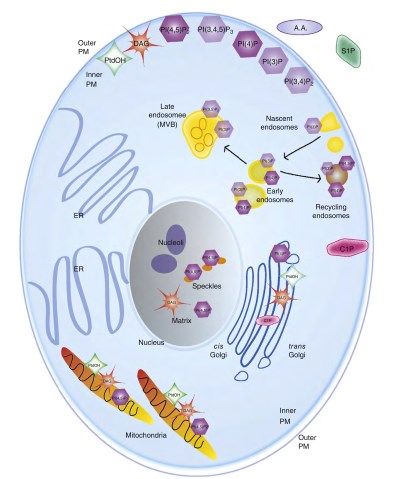
Figure 2. Spatial organization of signaling lipids. The subcellular locations of the major signaling lipids are represented in a stylized schematic of a mammalian cell. Note that all signaling lipids are present at the inner plasma membrane, but are not present at the outer plasma membrane. The most abundant phosphatidylinositols (by mass) have darker shading than the less abundant members.
Remodeling of PIPs results in enriched populations that are reliable markers for the various vesicle subtypes as they proceed through the endocytic cycle or split off into recycling endosomes (indicated by arrows).
Although DAG and PtdOH are abundant at the ER (the main site of phospholipid synthesis), these pools do not appear to be involved in signaling reactions and therefore are not represented in the diagram. DAG, diacylglycerol; ER, endoplasmic reticulum; MVB, multivesicular body; PtdOH, phosphatidic acid
While the temporal and spatial resolution of signaling lipids is still poor due to the high mobility, rapid metabolism, and low abundance of bioactive lipids within a cell, the combined data from fluorescent lipid-binding probes and lipidomic studies of organelles indicate that the distribution of signaling lipids varies throughout the cell. These observed distributions typically correspond with the functional requirements of the cell.
For example, trafficking pathways are highly dependent on PtdOH for fusion and budding of vesicles from the Golgi, and PtdOH can be seen concentrated at the Golgi apparatus. Interestingly, individual phosphatidylinositol subclasses are also enriched in various organelles, and control the recruitment of stage-specific proteins to different compartments.
For example, PI(4,5)P2 is highly concentrated in the cytosolic leaflet of the plasma membrane where it participates in the production of second messengers and is instrumental in endocytosis, PI(3)P is associated with early endosomes where it recruits adapter proteins, and PI(4)P is abundant at the Golgi apparatus where it is important for trafficking (Di Paolo and De Camilli, 2006; Mayinger, 2012).
Cellular partitioning is also evident for other lipids that serve structural roles, which is beyond the scope of this article but is outlined in several excellent reviews (Bader et al., 2011; Nicolson, 2014; Coskun and Simons, 2011).
Signaling lipids have recently been examined at the mitochondria, leading to several unexpected findings. Approximately 5% of the cellular PI(4,5)P2 is found at the mitochondrial outer membrane. While this seems like a minor amount, when normalized to the membrane surface area, the resulting density is approximately 25% of that found at the plasma membrane (Nemoto and De Camilli, 1999).
This pool of PIP2 has been shown to recruit a form of synaptojanin 2, a phosphatidylinositol phosphatase important for uncoating vesicles, to mitochondria, and appears to be important for the intracellular localization of mitochondria.
The phosphatidyli- nositol-4-phosphate 5-kinases which generate PI(4,5)P2 and a form of PLD that produces the substrate lipid for PIP2 production have also been identified at the mitochondrial membrane, indicating a separate PIP2 signaling network at this organelle. Subsequent studies have shown this network regulates fusion, fission, and localization of mitochondria in cells (Huang and Frohman, 2009).
Another important finding resulting from combined research efforts is the identification of independent lipid signaling pathways within the nuclear matrix. This finding was controversial for many years due to lack of evidence for any membranous structures in the nuclear matrix.
However, independent laboratories identified all the necessary components for a nuclear PI cycle, as well as nuclear lipid effectors such as the Star-PAP polyA polymerase (Li et al., 2013), PIP2- activated splicing factors, and nuclear DAG-binding proteins such as PKCa.
PI(4,5)P2 has been detected in situ using antibodies (Tabellini et al., 2003), and lipid microdomains (Cas- cianelli et al., 2008) and lipid droplets (Layerenza et al., 2013) have been detected in the nucleus. The combined evidence has resulted in the general acceptance of a separate lipid signaling web in the nucleus of cells.
Date added: 2024-06-13; views: 438;
