Applications in Molecular Biology and Gene Analysis
PCR has been adapted for a variety of uses in the laboratory in order to engineer DNA and to study gene expression. The three most common techniques, which will be discussed, are (1) the introduction of restriction sites into DNA, (2) the introduction of mutations into DNA, and (3) the analysis of gene expression using reverse transcriptase PCR (RT-PCR).
In order to amplify or clone a portion of DNA, the fragment must be introduced or ligated into a vector, such as a plasmid, for propagation. DNA products must be subcloned into sequences called restriction sites. Restriction sites are specific DNA sequences that are cleaved by enzymes. In order to subclone a DNA region of interest, the restriction site on the fragment of interest must match that of the vector.
The lack of convenient compatible restriction sites is often a problem which can make cloning difficult. PCR can be used to introduce restriction sites on the end of a DNA fragment of interest. The method used to accomplish this is summarized in Fig. 3. Oligonucleotide primers are designed that have the desired restriction sites in their 5' regions.
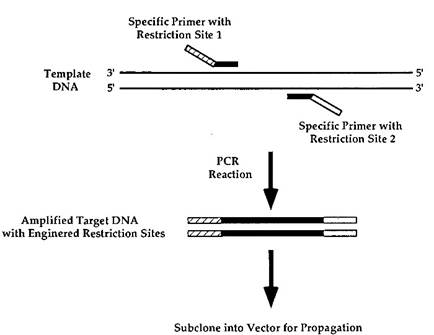
FIGURE 3. Molecular engineering of restriction sites onto a DNA fragment. The doublestranded DNA template is indicated. The primers shown contain two features. The regions indicated in black are homologous to the DNA target sequence of interest. The regions indicated by either the striped or the stippled boxes encode unique restriction sites that are not homologous to the DNA template. The PCR reaction is carried out, and an amplified product is obtained that contains the DNA region of interest flanked by restriction sites that can be utilized for cloning
Their 3' regions contain sequences that are specifically homologous to the region of interest. The PCR reaction is carried out whereby the presence of nonhomologous sequences in the primers will not affect their ability to hybridize to their target sequences. The resulting PCR product contains newly introduced restriction sites that can be utilized for cloning.
A second useful technique is the use of PCR for the introduction of mutations into DNA. The method used to do this is summarized in Fig. 4. Two PCR reactions are carried out separately. Each reaction is performed with one outer completely homologous primer and an internal primer that contains the mutation of interest.
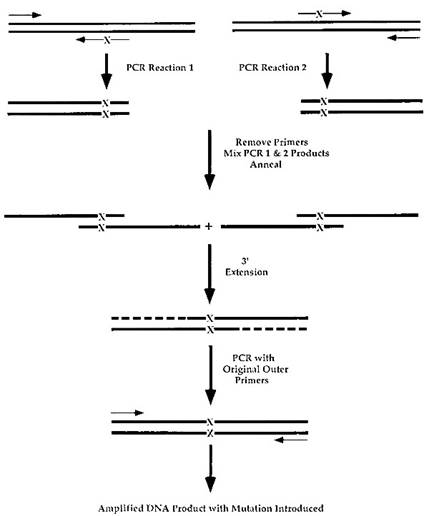
FIGURE 4. Introduction of mutations into a DNA target via PCR. The double-stranded DNA target in which a mutation will be introduced is indicated by the two heavy dark lines. The primers for amplification of the sequence are indicated by light lines with arrows. The nucleotide(s) that will be mutated is located in the internal primers and is indicated by an X. Initially, two PCR reactions are carried out to generate two smaller overlapping double-stranded DNA products that contain the mutation of interest. The products are purified, heat denatured, and allowed to reanneal. Overlapping DNA fragments are extended using polymerase to generate a double-stranded, full-length product containing the mutation. This product is subjected to PCR using the two original outer primers to amplify the target and yield a DNA fragment with a newly engineered mutation
This mutation can be a single or multiple nucleotide change as long as the primer contains enough completely homologous sequence to recognize its target region. Following the two PCR reactions, the samples are purified, allowed to anneal to each other, and the recessed 3' ends filled in enzymatically. The resultant molecule can then be amplified with the original two outer primers to generate a DNA fragment with a specific mutation introduced.
The introduction of mutations into DNA is useful for many reasons. One example is the study of protein function. A single amino acid of a protein can be examined by mutating it and analyzing the effect of this on protein activity. To mutate an amino acid, the three nucleotides encoding its corresponding codon in the DNA sequence are altered. The new protein sequence with the alteration can then be expressed and characterized. In addition to the introduction of mutations, PCR can also be used to insert and delete sequences from a DNA fragment of interest.
A third common use of PCR in the laboratory is in the study of gene transcriptional regulation. It is often of interest to examine whether the level of RNA transcribed from a particular gene is upregulated or downregulated over time or in response to specific reagents. However, sometimes the amount of RNA being examined is insufficient to be detected by typical methods. Amplifying a sequence from a small amount of RNA is called reverse transcriptase PCR or RT-PCR and is summarized in Fig. 5.
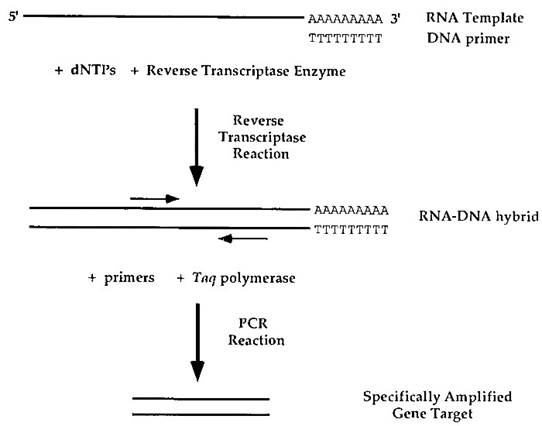
FIGURE 5. Reverse transcriptase polymerase chain reaction. An mRNA template is shown containing a polyadenylated 3' tail. A DNA oligonucleotide primer is hybridized to the RNA. A polythymidine DNA primer is shown; however, a gene-specific primer or a collection of random hexamers can also be used. The reverse transcriptase reaction is carried out to generate a DNA template as part of a DNA-RNA hybrid. Gene-specific primers and Taq polymerase are then added and a PCR reaction is carried out. The product can be analyzed by electrophoresis and quantitated to determine the relative transcription levels of the original RNA
The RNA of interest is first subjected to a reverse transcription reaction. Reverse transcriptase is a polymerase encoded by viruses that is able to synthesize a DNA strand from a RNA strand by extension of a DNA primer. Several types of DNA primers can be used in the reverse transcriptase reaction. One type is a specific primer that is homologous to the gene of interest. A second type, illustrated here, is a polythymidine oligonucleotide that can hybridize to the polyadenine sequence that is present at the 3' end of eukaryotic mRNAs.
A third type is a mixture of random hexamer oligonucleotides that can hybridize and therefore, theoretically, prime at all RNA sequences present, thus converting all RNAs to DNAs. Following this first-strand DNA synthesis from RNA, gene-specific primers are utilized and a typical PCR reaction is carried out. Samples can be analyzed by electrophoresis and compared to determine the relative amounts of original RNA that was present.
Date added: 2024-07-02; views: 565;
