Polymerase Chain Reaction
The polymerase chain reaction (pcr), more than any other discovery in recent history, has revolutionized both basic medical research and the clinical diagnosis of disease. The PCR reaction allows multiple rounds of amplification of a minimal amount of starting DNA material to obtain sufficient quantities of sample for analysis and use. The exponential amplification of DNA is facilitated by the use of a heat- stable DNA polymerase, isolated from a thermophilic bacterial organism.
The use of this thermostable polymerase is essential to the reaction because each amplification cycle requires a high temperature DNA denaturing step. Since its first description in 1985 as a means to amplify DNA, PCR methodology has been modified and improved so that it is now utilized for many applications beyond this original purpose.
In biomedical research, PCR is used to introduce mutations into genes to study protein-structure function, analyze gene transcriptional regulation, and facilitate cloning. In paleontology, PCR is utilized to amplify and characterize fossil DNA. In forensic analysis, PCR is utilized to analyze minimal quantities of tissue evidence that can be used as a “fingerprint” in criminal identification.
Introduction. The PCR reaction is an extension of work originally done in defining the function of a class of enzymes called polymerases. Polymerases are enzymes that are able to catalyze a reaction in which a new strand of DNA is synthesized using a preexisting strand of DNA as a template. The steps of this reaction are seen in Fig. 1.
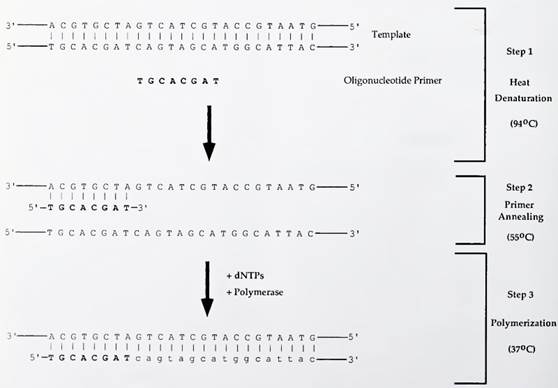
FIGURE I. Steps of a standard polymerization reaction. The capital regular type letters indicate nucleotides of the double-stranded DNA template and the capital bold type letters indicate the primer. The reaction is heated to melt or denature the double-stranded DNA, and the oligonucleotide primer is then allowed to anneal to the now single-stranded template. Polymerase and free deoxynucleoside triphosphates (dNTPs) are added. The enzyme extends the primer by the addition of dNTPs to synthesize a new DNA strand complementary to the template. The newly synthesized DNA strand is indicated by lowercase regular type letters. The temperatures indicated are typical ones for a polymerization reaction
In step 1, a double-stranded piece of DNA, the template, is mixed with another short oligonucleotide fragment of DNA, called a primer, that is complementary to a segment of the long template strand. The mixture is heated, typically at 94°C, in order to “melt” or dissociate the two DNA template strands. The mixture is then cooled to allow the primer sequence to anneal to the template (Fig. 1, step 2).
The annealing temperature is typically 55°C; however, this can vary depending on the degree of homology between the primer and the template. The polymerase enzyme and a mixture of the four deoxynucleoside triphosphates (dNTPs: dATP, dCTP, dGTP, and dTTP) are added to the reaction (Fig. 1, step 3). Because typical polymerases are thermolabile, this addition is done following step 1 and step 2 as the high temperature of these previous steps can cause denaturation of the protein.
The reaction is then incubated, allowing the polymerase to sequentially add free nucleotides in a 5' to 3' direction to the growing primer until a new DNA strand, complementary to the original template, is synthesized. Kary Mullis, who subsequently won the Nobel Prize for his work, first demonstrated that this reaction could be repeated or cycled multiple times in order to amplify a DNA target template over a millionfold.
Date added: 2024-07-02; views: 498;
