Polymerase Chain Reaction (PCR): Theory and Methodology
The cycling or chaining of a single polymerase reaction to amplify a DNA template is driven by several features that are illustrated in Fig. 2. In the first step of a PCR reaction, two different oligonucleotide primers are used which flank the region of DNA to be amplified.
The polymerase reaction is carried out as described earlier, and a single cycle will thus generate two new complementary DNA strands. These complementary strands are part of two new double-stranded DNA products that can be utilized in the next reaction cycle as template. Thus, each subsequent reaction step doubles the amount of the new double-stranded product generated.
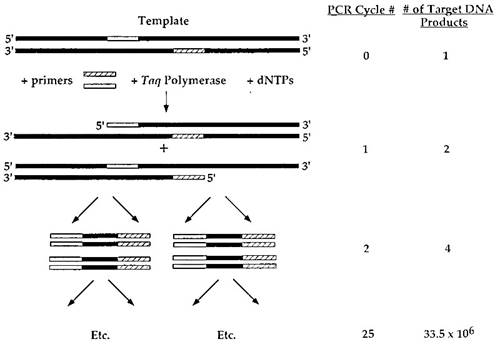
FIGURE 2. The polymerase chain reaction. The double-stranded DNA template is indicated in bold. Both primers and regions of the DNA template that are homologous to the primers are indicated by stippled or striped boxes. On the right, the PCR cycle number and the corresponding number of molecular DNA products that would be synthesized at that cycle number are indicated. At each step, the amount of product is effectively doubled. A PCR reaction is typically carried out for 25 cycles and can therefore give a 33.5-millionfold amplification of starting material
The ability to synthesize a new product at each step is driven by the addition of a large molar excess of the oligonucleotide primers added at the first cycle. Therefore, sufficient primer is available for use at each step to hybridize to the newly formed template and to initiate each subsequent reaction. PCR reactions are typically carried out for 25 cycles. Because the amount of product template generated is doubled at each step, the amount of amplification of the initial template is 225 or 33.5 millionfold.
The practical use of PCR as a laboratory tool was initially hampered by two problems: (1) because different temperatures are used for the melting, annealing, and polymerization steps, samples had to be manually transferred to different water baths during the entire cycling amplification procedure and (2) the polymerase utilized for the reaction, Klenow enzyme derived from the bacteria Escherichia coli, was heat labile. Therefore, after each cycle, it was necessary to add new Klenow enzyme to the samples.
Since 25 cycles of a PCR reaction take several hours to complete, the amplification process was a technically tedious procedure. The first problem was overcome by the development of a computerized cycling machine that could be programmed to alter the temperature of a heating block to the preset PCR parameters. The second problem was solved with the discovery and isolation of a heat-stable enzyme, called Taq polymerase, that could carry out the reaction.
Taq polymerase was cloned from a bacterium called Thermus aquaticus, which was first described in 1969. This thermophilic microorganism was isolated from a hot spring in Yellowstone National Park and is capable of growth at 70-75°C. Because of its adaptation for growth in this environment, the DNA polymerase expressed by T. aquaticus carries out its polymerization reaction at high temperatures (70°C) and is relatively stable and not denatured by exposure to even higher temperatures (90°C).
The heat stability of Taq polymerase makes it highly suitable for use in PCR. Taq can be added to a PCR reaction at the first step and can be utilized without replenishing throughout all subsequent cycles, including each 90°C melting step. The use of Taq coupled with the advent of automatic robotic cyclers has made PCR an efficient routine process.
Date added: 2024-07-02; views: 599;
