Acquired Immunodeficiency Syndrome (Virology)
Structure, Proteins, and Genes. Virion Structure. The AIDS virus carries its genetic information in two identical RNA molecules. Each RNA strand is approximately 9500 nucleotides long, roughly one- millionth the size of the genome of the Escherichia coli bacterium. A cylindrical core made up of viral protein encloses the genome and a few associated viral proteins, forming the nucleocapsid. This nu- cleoprotein core, in turn, is surrounded by a lipid membrane (i.e., the viral envelope) derived from the infected cell.
A layer of viral protein forms the backbone of the viral envelope, and approximately 72 large knoblike structures, composed of the viral glycoproteins (i.e., proteins with sugar residues), protrude through the envelope to the virion exterior. The complete virus is a spherical particle that averages 100 nm in diameter (Fig. 1).
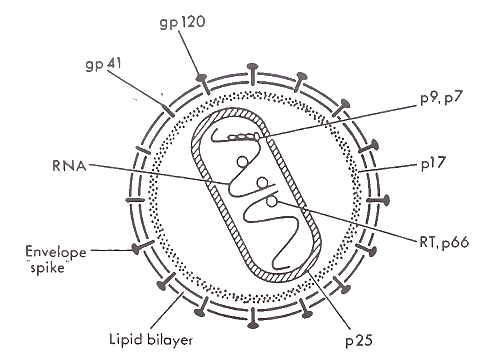
FIGURE 1. An HIV virion. From J. A. Levy (1989) with permission
Proteins and Genes. The envelope glycoprotein is the first HIV protein to interact with a cell. Because of this central role in initiating infection, this viral glycoprotein has been extensively studied. It consists of two subunits (gpl20 and gp41) with molecular weights of 120,000 and 41,000, respectively. gpl20 has approximately 480 amino acids and is heavily glycosylated; about 50% of its total molecular weight is due to sugar residues. The entire molecule lies outside the viral membrane, but remains attached to the virus by a noncovalent interaction with the gp41 membrane- spanning subunit.
gp41 has approximately 345 amino acids and is much less glycosylated. Near its carboxy-terminal end is an apparent membrane-spanning domain of approximately 22 amino acids. This region presumably anchors the envelope glycoprotein complex in the membrane, but the topology of the molecule in the membrane is not known. A short tail of approximately 150 amino acids extends beyond the membrane inside the virus. A region near the amino- terminal end of gp41 appears to interact with gpl20, but the interaction is relatively weak. gpl20 can easily dissociate from the gp41 subunit and can be found in the sera of infected individuals and in supernatants of infected cultures.
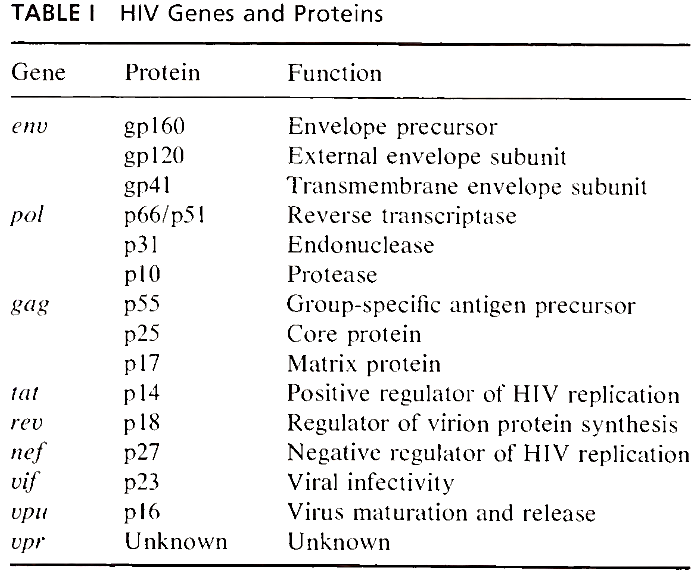
gpl20 and gp41 are both products of the env gene, located near the downstream (i.e., 3') end of the viral genome (Fig. 2 and Table I). They are synthesized as a polyprotein precursor of 160 kDa and cleaved into two linked subunits as the maturing protein is being transported from the cytoplasm of the cell, where it is made, to its final destination at the cell surface. The cleavage is required in order for HIV to be infectious and is performed by a cellular enzyme. Thus, cells lacking this enzyme cannot perpetuate HIV infection. Such bipartite envelope glycoproteins derived from polyprotein precursors are a highly conserved characteristic among different classes of enveloped viruses.
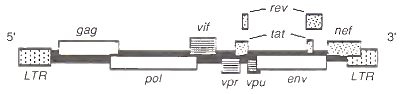
FIGURE 2. An HIV-1 genome, as, integrated in the host genome
The three-dimensional structure of the envelope glycoprotein complex is not known, but it is thought that three or four gpl20-gp41 molecules join to form the knoblike spikes seen in electron micrographs. The three-dimensional conformation of gpl20, including its sugar residues, seems to be functionally important, because several spatially distinct regions in the carboxy-terminal end of gpl20 appear to be involved in binding HIV to the CD4 cellular receptor (see Section II,A).
In addition to serving as an anchor for the gpl20 subunits, gp41 appears to play a critical role in allowing the virus to penetrate and fuse with the target cell membrane. Because of its striking similarity to documented fusion-inducing sequences of other viruses, a hydrophobic stretch of about 30 amino acids, beginning at the amino terminus (where gp41 is cleaved from gp 120), is thought to fuse the virus to its target cell. This domain is also one of the more highly conserved regions of the HIV env gene, which is otherwise variable.
Beneath the envelope are the matrix lining and the nucleoprotein core, both made up of the group- specific antigens. These gag proteins are so named because the amino acid sequences of these proteins, unlike the envelope glycoproteins, are relatively invariant among different viral strains. All gag proteins are encoded by the gag gene, located near the beginning (i.e., the 5' end) of the viral genome.
The gag proteins are derived from a 55-kDa polyprotein precursor that is cleaved by an HIV-encoded protease into several smaller proteins (i.e., p25, p 17, p9, and p7). p 17 attaches to the viral membrane via a fatty acid myristyl group and extends into the interior of the virus, an unusual location for a retroviral gag protein. pl7 then forms a matrix that could be important in maintaining the shape of the viral particle. p25 is the major component of the viral nucleoprotein core and is the most abundant HIV protein. The smaller gag proteins can be found within the core, but their functions are not well defined.
Associated with the diploid viral genome are three viral enzymes encoded by the pol gene, which overlaps the gag gene (Fig. 2). Reverse transcriptase copies the viral RNA into DNA, the reverse of the usual flow of genetic information from DNA to RNA. Once copied into double-stranded DNA, in- tegrase (i.e., endonuclease) helps insert the viral DNA into the host cell genome. Finally, protease is involved in processing viral proteins as they form mature viral particles. All pol proteins are derived from a gag-pol fusion polyprotein, which results when the ribosome infrequently shifts its reading frame at the gag-pol junction and translates the pol transcript. The fusion polyprotein is later cleaved into its components by protease as the progeny virions are released from the cell (see Section II,D).
So far, the description of the HIV structural genes (i.e., genes for proteins that are present in the mature virion) suggest that HIV is a “straightforward” retrovirus. However, in addition to the gag, pol, and env genes present in all retroviruses, HIV has at least six other genes (Fig. 2). Many of these genes code for proteins that regulate HIV expression, suggesting a retrovirus of unusual complexity. Nine genes have been identified. Two are divided, many overlap, and several appear to be novel genes without direct counterparts in other retroviruses.
The tat and rev genes have protein-coding sequences that are divided into two segments. Their mRNAs are derived from RNA splicing of precursor transcripts that contain intervening noncoding sequences. Both tat and rev are indispensable for generating large amounts of virus particles, yet their modes of action are very different.
The tat gene gets its name from the ability of its protein product to activate in trans. trans-Activation is an experimentally defined phenomenon. It means that the gene product, usually a soluble protein, is able to activate genes located on a different chromosome, instead of being restricted to genes located nearby on the same chromosome. Thus, tat proteins can potentially activate genes in many places in the genome. However, only genes that have a specific responsive element to tat can be activated by the tat protein, regardless of the location.
Responsive elements are short stretches of nucleotides usually located near the start of the proteincoding part of the gene. Because the responsive elements are located on the same chromosome as the genes they affect, they are said to act in cis. The responsive element for tat is TAR, for trans-acting responsive sequence. It is located at the 5' end of the viral genome in the long terminal repeat (LTR) region (see Section II,C) and is present in HIV mRNA.
The 14-kDa tat protein has a dramatic effect on virus production. In its absence HIV is made inefficiently, if at all. When present, the tat protein increases the expression of all HIV genes, including tat, by several orders of magnitude. Such a positive-feedback loop can give rise to massive amounts of viral progeny.
The mechanism of tat activation has not been clearly defined, but currently experimental evidence suggest that tat increases both transcription of viral genes and translation of viral message, especially the former. These activities appear to involve the TAR element, which when present as singlestranded mRNA, is predicted to form a hairpin-type loop structure. Experiments involving mutations that affect the secondary structure of TAR have shown that the hairpin-type structure is necessary for tat regulation. However it is unclear yet whether the tat protein binds these sequences directly or modifies other cellular proteins that recognize it.
The rev protein, in contrast, appears to be important in regulating which mRNAs are available for translation into protein. Without the rev protein the mRNAs for the gag, pol, and env structural proteins are not available to the translational machinery in the cytoplasm, yet the short multiply spliced mRNAs for the regulatory proteins such as tat and rev are translated. Mutant viruses that lack a functional rev protein are therefore unable to generate progeny virions. The apparent block of cytoplasmic transport of the structural mRNAs, as and as well as the genomic.RNA, is due to a cA-acting repression element (CRS) present in the gag and env sequences. These sequences are spliced out of the shorter mRNAs such as those for tat and rev.
The rev protein overcomes the negative effects of CRS, allowing accumulation of CRS-containing mRNAs in the cytoplasm by interacting with a different element in the mRNA called cA-acting antirepression element (CAR, also known as RRE for reu-responsive element). The CAR element in the env mRNA is predicted to form a complex secondary structure involving multiple stem loops. By specifically binding to CAR, the rev protein apparently allows the CRS-containing mRNA to be transported to the cytoplasm. The mechanism is not clear, but there is some evidence to suggest that splicing complexes are involved.
The control of HIV gene expression may be further influenced by a third viral protein encoded by the nef gene, located at the 3' end of the viral genome (Fig. 2). The function of this nonessential protein is not known, although some experiments indicate that it may down-regulate the growth of some HIV strains. Other clues to the nef protein function come from studies of its biochemical properties. The nef protein is a 27 kDa phosphoprotein with a fatty acid myristyl group covalently linked to its N-terminus.
This fatty acid group probably anchors the nef protein to the cytoplasmic leaflet of the plasma membrane. When produced by genetically engineered bacteria, the nef protein exhibits GTPase, autophosphorylation, and GTP-binding properties. The location and biochemical properties of the nef protein suggest that it could play a role in signal transduction or cellular metabolism. Interestingly, these properties are also shared by proteins of the ras oncogene family.
The vif gene (for virus infectivity factor) codes for a 23-kDa basic protein that appears to play a role in assuring the infectivity of virus particles. Mutations in the vif gene result in viruses that have a reduced ability to infect cells, even though the viruses appear normal in other respects. Nevertheless, cells infected by u/f-mutant viruses seem to be able to transmit HIV to uninfected cells as efficiently as cells infected with viruses that lack mutations in vif presumably via some cell-to-cell mechanism (see Section III,A,1). The vif protein is found primarily in the cytoplasm of infected cells, but may also be present in virus particles.
The vpu gene codes for a 16 kDa protein that appears to be involved in the release of progeny virions from infected cells. The vpu protein is phos- phorylated and probably anchored in a membrane by a hydrophobic stretch of amino acids at its N terminus. Yet, despite being an integral membrane protein, the vpu protein has never been detected in virions, and indirect immunofluorescent microscopy shows that the vpu protein is concentrated in perinuclear regions, most likely in the Golgi complex.
Cells infected with viruses that do not make the vpu protein show an increased intracellular accumulation of viral proteins and a delayed and decreased production of progeny virions in culture supernatants. In addition, the upu-mutant viruses show aberrant budding; core sizes vary, are abnormal, and assemble at intracytoplasmic membranes. These findings suggest that the vpu protein plays a role in virion maturation and release.
The function of the vpr protein is less well defined. Although the vpr gene is generally well conserved among HIV strains, no function has been associated with the 78 amino acid vpr protein. Mutational analyses indicate the protein is clearly not essential for viral replication, though one study found that cells infected with low titers of upr-mutant virus show a delay in virus production.
Date added: 2023-05-09; views: 647;
