During transcription: A secret cipher for transcribing DNA to RNA
After discovering DNA’s structured in the 1950s, the next great mystery was how DNA could hold the information for other molecules such as RNA and proteins. It took decades of research to unravel this.
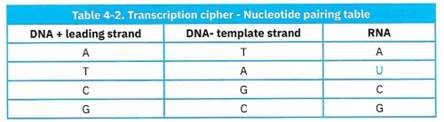
As you saw in Figure 4-19 and Figure 4-20, RNA nucleotide structures are quite similar to DNA nucleotides. There is one other significant difference between DNA and RNA: There is no ‘T’ thymidine nucleotide in RNA. Instead, there is a TJ’ nucleotide for uridine, hence, GCAU.
In Chapter 1, you saw that certain nucleotides can complement each other using Chargaff’s Rule. Notably, A’s can bind to T’s to form a double-stranded DNA, and C’s can bind to G’s. This complementary rule is how RNA polymerase knows which ribonucleotides to match up with when transcribing an RNA strand, and this is why it always reads the template strand. It uses the template strand because it is a ‘mirror image’ of the leading (+) strand. The end goal of RNA polymerase is to have an RNA strand that is a replica of the leading (+) DNA sequence, making the (-) template strand its ‘mirrored strand’.
Lastly, because there is no T in RNA, T’s are replaced with U’s, (Table 4-2). This means that the ‘A’ in a (-) template strand interacts with a ‘U’ instead of a ‘T\ In other words, if there is a‘T’ in your DNA (+) strand, then there will be a ‘U’ in the RNA strand.
As RNA polymerase moves downstream reading the template strand of the DNA, ribonucleotides doing the Four B’s bump into the RNA polymerase. When the correct complementary ribonucleotide (Table 4-2) bumps the correct DNA nucleotide inside the RNA polymerase (e.gA-U or G-C or T-A) it will bind and trigger a chemical reaction burst, permanently attaching itself to the growing string of RNA (Figure 4-29).
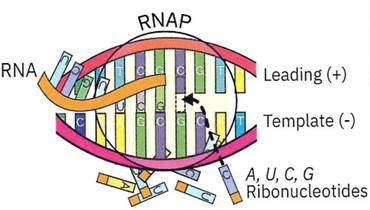
Figure 4-29. As RNA polymerase travels downstream unzipping the double-stranded DNA, it “reads” the (-) template strand. Free-floating, complimentary ribonucleotides (C-G or U-A or T-A) will bind strongly to the nucleotide of the DNA’s template strand within the polymerase. This triggers the polymerase to attach the ribonucleotide to the growing RNA strand. If a nucleotide doesn’t bind strongly (for example a C-A), it will bump out of the RNA polymerase. Eventually, the correct one will enter and be added to the growing string of
If an incorrect match occurs (e.g., A-G), then the ribonucleotide will bump out. Eventually, a correct ribonucleotide will bump in, bind strongly, and trigger the reaction. When this chemical reaction happens, it also propels the RNA polymerase to move further down the DNA strand to the next nucleotide. The RNA polymerase moves at about 50 ribonucleotides per second - that’s fast! Can you do the following exercise just as fast? Complete Table 4-3 in less than one second!
Date added: 2023-11-02; views: 654;
