Ultraviolet-B Perception and Signalling
Many transcriptional responses to increased doses of UV-B are non-specific and shared with, for instance, defence responses against pathogen attack (Stratmann 2003). The respective signalling has been associated with UV-B as a severe stressor of plant life, is triggered by DNA damage and ROS production, and involves typical plant stress hormones (Fig. 3.29).
Responses result in repair and in protection by the same classes of chemical compounds, which can act either as sunscreen pigments or as components for defence (phytoalexins). In particular, induction or reinforcement of radical scavenging systems such as glutathione reductase, ascorbate peroxidase and glutathione peroxidase are very important for avoidance and alleviation of UV-triggered stress. Overall, responses to UV-B stress are dependent on signalling pathways triggered by non-specific secondary stress, while photomorphogenic acclimation responses are more UV-B specific and are triggered by low levels of UV. It is important to note that both contribute to survival.
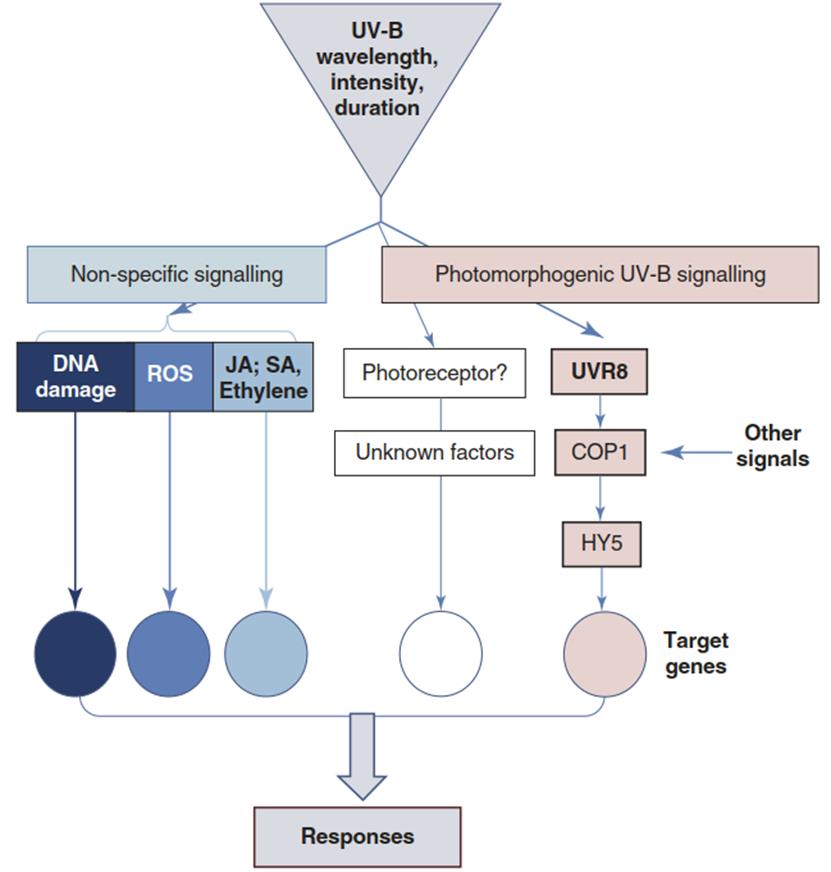
Fig. 3.29. Ultraviolet (UV)-B signal transduction pathways. UV-B induces UV-B-specific and non-specific signal production and transduction. The UV-B wavelength, intensity and duration of exposition trigger the induction of specific sets of target genes and downstream responses that result in repair of UV-triggered damage and adaptation to UV-B. JA jasmonic acid, SA salicylic acid. (Modified after Jenkins (2009))
The specific responses can—depending on the wavelength, intensity or duration—also represent acclimation and avoidance reactions. During the past decade, much progress has been made in the elucidation of signal transduction chains resulting in UV-B tolerance of plants.
Photomorphogenic signalling requires perception of radiation. While the photoreceptors for visible light contain chromophores, the UV-B photoreceptor lacks such a component. It absorbs shortwave radiation by a series of aromatic amino acid residues—in particular, 14 tryptophan residues (Trp, W). The UV-B receptor was originally described as a regulatory protein in UV-triggered signal transduction in A. thaliana (Kliebenstein et al. 2002) and was termed UV RESISTANCE LOCUS 8 (UVR8).
In 2011, Rizzini et al. identified this protein as the long- sought-after UV-B photoreceptor which, by a special arrangement of tryptophan residues and positively charged arginine residues, can convert UV-B radiation into a chemical signal. Other authors contributed the detailed photochemical and biochemical mechanisms of that process. In all investigated species, from algae to higher plants, this type of UV-B photoreceptor has been found (Rizzini et al. 2011).
In the energetic ground state (e.g. in the dark), UVR8 is a doughnut-shaped homodimer whose monomers are linked by a network of salt bridges and aromatic side-chain interactions (Gardner and Correa 2012) (Fig. 3.30). Each monomer consists of 440 amino acid residues with 14 Trp residues, which are clustered at the top surface where the dimer forms. Three of them form a triad, which interacts with another tryptophan of the counterpart analogue (Fig. 3.30).
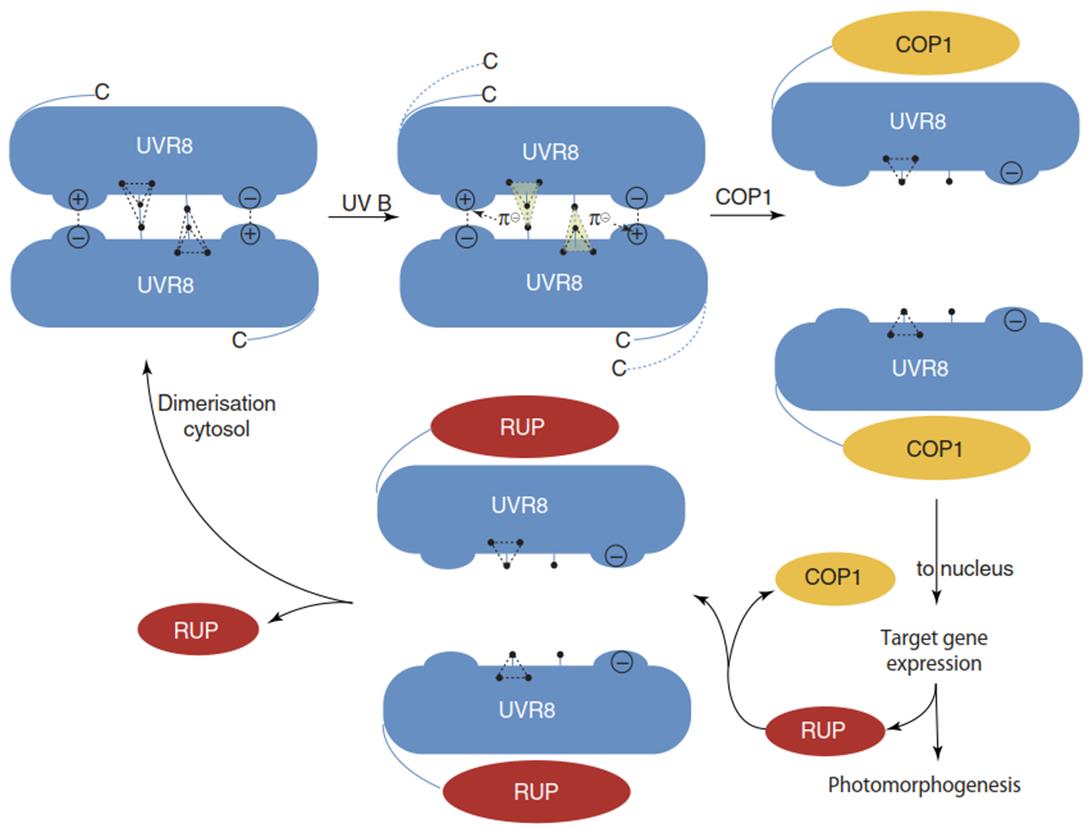
Fig. 3.30. Plant ultraviolet (UV)-B sensing by the UVR8 photoreceptor. UVR8 exists as a stable dimer in the dark through salt bridges and interactions of tryptophan residues forming a Trp-pyramid (3 + 1 trp residues •). Excitation of the tryptophan residues by UV-B results in the donation of n electrons to the arginine residues of the salt bridges, leading to charge neutralisation and dissociation of the dimer. Monomeric UVR8 binds to COP 1 which, as a complex, can migrate into the nucleus for expression of the HY5 transcription factor and other genes that lead to the formation of, for example, UV screens. Among these genes is that for RUP proteins (REPRESSOR OF UV-B PHOTOMORPHOGENESIS), which can replace COP1 and finally, by dissociation from UVR8, facilitate dimerisation of (inactive) UVR8. (Modified from Gardner and Correa (2012); for details, see Jenkins (2014))
Excitation of the tryptophan residues by UV-B results in the dissociation of the salt bridges and releases the monomers. The active UVR8 monomer then binds to the multifunctional COP1 protein, which is a central regulator in UV-B and visible light signalling (Oravecz et al. 2006) (Fig. 3.22). At this point the question of “normal photomorphogenesis” versus the UV stress response arises again but cannot be conclusively answered. It is, however, clear that UV-B-specific responses— whether they are contributing to the normal development of a plant or are excited by an unusual dose of UV radiation—are mediated by the UVR8-COP1 signalling pathway (Heijde and Ulm 2012).
The central protein of this pathway is COP1, best known as a negative regulator or repressor of photomorphogenesis (Fig. 3.22). However, in UV-B signalling, COP1 is a positive regulator. The interaction complex of monomeric UVR8 with COP1 migrates from the cytosol into the nucleus, where it activates HY5 gene expression. The transcription factor HY5 triggers the expression of a great variety of UV-B-responsive genes. Among these are genes that encode proteins required for UV-B tolerance or protection, such as photolyases for DNA repair, and enzymes of the phenylpropanoid pathway, such as chalcone synthase, by which phenolic UV-B scavengers are produced.
References:
Ahn TK, Avenson TJ, Ballottari M, Cheng Y-C, Niyogi KK, Bassi R, Fleming GR (2008) Architecture of a charge-transfer state regulating light harvesting in a plant antenna protein. Science 320:794-797
Anderson JM, Andersson B (1988) The dynamic photosynthetic membrane and regulation of solar energy conversion. Trends Biochem Sci 13:351-355
Barnes JD, Percy KE, Paul ND, Jones P, McLaughlin CK, Mullineaux PM, Creissen G, Wellburn AR (1996) The influence of UV-B radiation on the physicochemical nature of tobacco (Nicotiana tabacum L.) leaf surfaces. J Exp Bot 47:99-109
Berg JM, Tymoczko JL, Gatto Jr. CJ, Stryer L (2015) Biochemistry, Internatl edn. Springer, Berlin, Heidelberg
Bjorkman O, Demming-Adams B (1994) Regulation of photosynthetic light capture, conversion, and dissipation in leaves of higher plants, Ecol Stud. Springer, Heidelberg, New York; 100:17-47.
Bjorkman O, Powles SB (1981) Leaf movement in the shade species Oxalis oregana. I. Response to light level and quality. Carnegie Inst Wash Year B 80:59-62
Buchanan BB, Gruissem W, Jones RL (2015) Biochemistry and molecular biology of plants, 2nd edn. Wiley, Hoboken
Casal JJ (2013) Photoreceptor signaling networks in plant responses to shade. Annu Rev Plant Biol 64:403-427
Cashmore AR, Jarillo JA, Wu Y-J, Liu D (1999) Cryptochromes: blue light receptors for plants and animals. Science 284:760-765
Chelle M, Evers JB, Combes D, Varlet-Grancher C, Vos J, Andrieu B (2007) Simulation of the three-dimensional distribution of the red:far-red ratio within crop canopies. New Phytol 176:223-234
Date added: 2025-01-17; views: 413;
