Comparison of ТЕМ and FF on the same cell type. Diagram of a nucleus showing the nuclear envelope
Fig. 1-7. Freeze fracture of two adjacent cells. The extracellular space passes diagonally from middle left to upper right, separating the particle-rich PF face of the top cell from the particle- poor EF face of the bottom cell at lower left. At mid-right, note the approximation of both leaflets (EF and PF) of the lower cell plasmalemma, as seen in cross-fracture, (x 70,000.).


Fig. 1-8. Comparison of ТЕМ and FF on the same cell type. A, ТЕМ of a cell shows nucleus (N), mitochondria (M), lysosomes (L), and cell surface (plasmalemma). (x 17,000.) B, FF of a cell with nucleus (N) and nuclear pores, lysosomes, endoplasmic reticulum (er), and the protoplasmic face of the cell plasmalemma (CC). (x 3,000.).
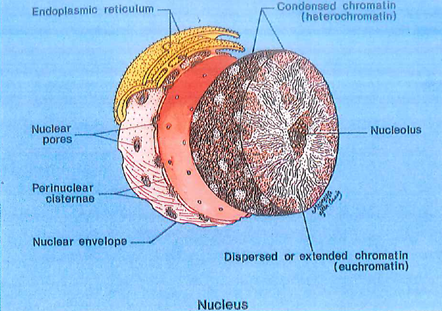
Fig. 1-9. Diagram of a nucleus showing the nuclear envelope, a double membrane structure with pores through which small molecules may pass between the cytoplasm and the nucleus. Cisternae of the endoplasmic reticulum may be directly connected to the nuclear envelope. Heterochromatin (inactive) is associated with the nuclear membrane, while euchromatin is actively involved with formation of RNA.
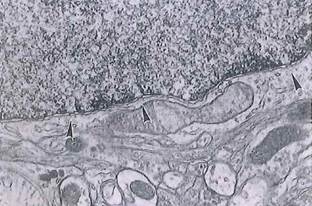
Fig. 1-10. ТЕМ of a cell revealing diaphragm-covered pores (arrowheads) in the nuclear envelope. Note the difference between euchromatin and heterochromatin. (x 29,000.).
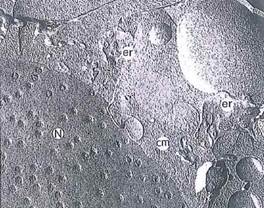
Fig. 1-11. Freeze fracture revealing pores in an otherwise smooth nuclear envelope. Surrounding the nucleus are the cytoplasmic matrix (cm) and elements of endoplasmic reticulum (er). Direction of the platinum shadowing is from upper right in this FF. (X 32,900.).
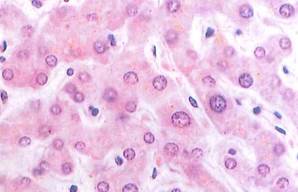
Fig. 1-13. LM of the liver. Note the large, vesicular nuclei of the liver cells, as well as the dark, more condensed nuclei of cells lining (or within) the vascular channels that separate rows of liver cells. This stain does not reveal the numerous mitochondria present within these cells. Compare this image with that in Fig. 1-16. (H&E; X450.).
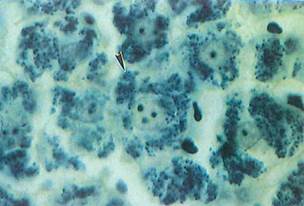
Fig. 1-14. LM of liver that has been stained with iron hematoxylin to demonstrate mitochondria (arrowhead) within the cytoplasm. The amorphous shapes in the vascular channels between liver cells are heavily stained red blood cells. (Iron hematoxylin; X750.).
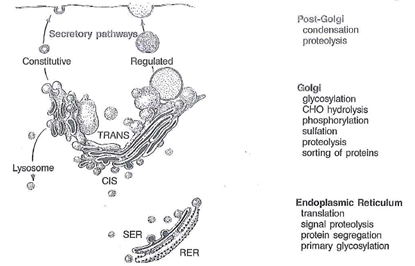
Fig. 1-16. Schematic drawing of the Golgi apparatus depicting the relationships between the synthesis of proteins in the rough endoplasmic reticulum (RER) and the functional compartments traversed until these secretory proteins are released by exocytosis. The constitutive pathway is nonregulated and consists of dilute packing of proteins that are continuously discharged from the cell. The regulated pathway utilizes storage of product in secretion granules that are released on stimulation by secretogogues.
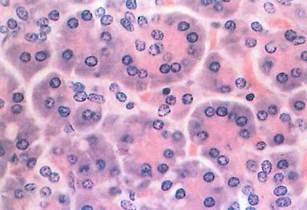
Fig. 1-17. High-magnification LM revealing the structure of the pancreatic acinus. Nuclei are located near the base of the cells; the Golgi apparatus lies in a supranuclear position but is not visible with this stain. Compare this image with Fig. 1-18, which is also pancreas. (H&E; X350.).

Fig. 1-18. Profiles of Golgi apparatus seen by LM in cells of the pancreatic acini, lying between unstained nuclei and cell apices. Secretions of these cells are released through the apical membranes into the lumen of the acinus. (Silver impregnation and gold toning; X500.).
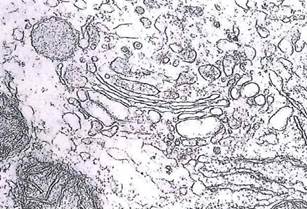
Fig. 1-19. ТЕМ of a liver cell showing a Golgi apparatus with three to five cisternae. The cis face of the cisternae is oriented toward the RER, while the dilated rims of the opposite trans face gives rise to vesicles/vacuoles that form secretion granules. (ХЗЗД00.).
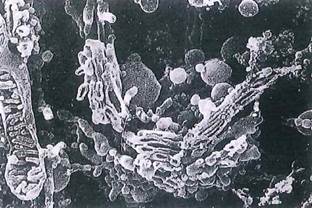
Fig. 1-20. SEM of the Golgi apparatus in an extracted cell, which reveals the spatial relationships between adjacent cisternae of the Golgi apparatus and forming vesicles. In the adjacent fractured mitochondrion, note the organization of cristae within the matrix (compare to Figs. 1-12 and 1-15). (x 6,300.).
Date added: 2022-12-11; views: 1074;
