Ice Nucleation Proteins
The so-called ice nucleation-active bacteria (INA bacteria) produce cylindrical surface proteins, which facilitate heterogeneous nucleation (ice nucleation proteins (INPs)). These bacteria are commonly gram-negative, epiphytic and pathogenic (Lorv et al. 2014). An example is Pseudomonas syringae. The effect of the “decoration” of the outer membrane with INPs is shown in Table 4.6 in comparison with other bacteria, which do not produce these proteins.
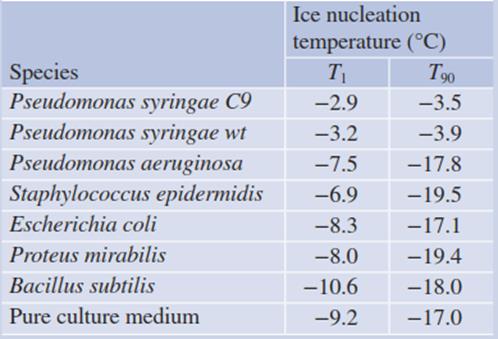
Table 4.6. Ice nucleation activity of bacterial cultures (Maki et al. 1974). Thirty droplets (each 0.01 mL) of test material were placed on a controlled surface, and the temperature was slowly lowered from room temperature to -25 °C. The temperature at which the first crystal formed (T1) and the temperature at which 90% of the droplets were frozen (T90) were recorded
The INP from P syringae consists of a particular octapeptide in up to more than 100-fold repetition (Fig. 4.18). The N terminus is anchored with transmembrane spans to a glycosylphosphatidylinositol patch of the outer membrane of the bacterium (Turner et al. 1991; Sarhan 2011).
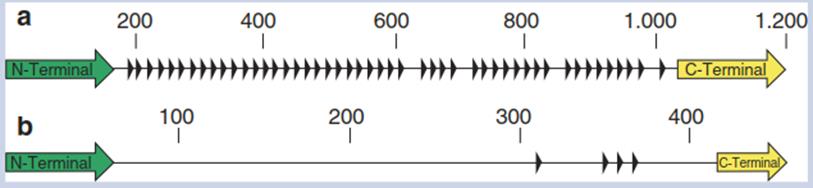
Fig. 4.18. Blueprints of the ice nucleation protein (INP) from Pseudomonas syringae a and INP-F from the fungus Fusarium acuminatum b. Triangles represent the repetitive motif. The N-terminal domain (about 15%) of the Pseudomonas INP consists of three or four potential transmembrane spans; the C-terminal tail makes up 4% of the protein (Shimazu et al. 2003). The smaller number of the IN motifs in the fungal protein result in lower ice nucleation activity: P syringae: —2 °C, F. acuminatum: —5 °C. (Lagzian et al. 2014)
Deletion mutants show that a 68-fold repetition of the motif is sufficient to effectively trigger ice seeding. A change in the octapeptide sequence abolishes ice nucleation activity (Green and Warren 1985). A model of the protein predicts a largely planar molecule with a molecular mass around 150 kDa, serving as a template for forcing water into an ice lattice. This quasi-crystalline fixation of the water molecules minimises the degree of supercooling that is required to “quiet” the water clusters (Kajava and Lindow 1993) (Table 4.7).
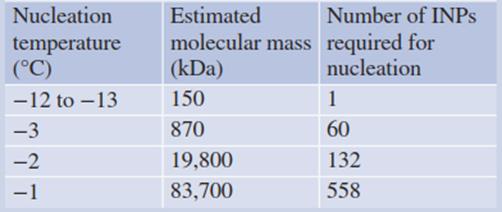
Table 4.7. Analysis of bacterial ice nucleators (INPs) from Pseudomonas syringae. (Hew and Yang 1992)
As mentioned above, ice seeding activity has been observed with plant cell walls from blueberry stems and also with cell walls fromrye leaves (Brush et al. 1994). Apart from a general chemical composition (proteins, carbohydrates and phospholipids (Gusta et al. 2004)), the mode of function and mechanism of plant ice nucleators are not yet known.
The effects of INA bacteria are double edged. In frost-tolerant plants they trigger “high-temperature” ice seeding, thus avoiding strong supercooling and potential intracellular ice formation. On the other hand, they cause frost damage to many crops, such as maize, strawberry and citrus fruits, which are not freezing resistant but can tolerate supercooling to some extent. Attempts have been made to dilute out the INA bacterial infestation of crop plants by replacement with antagonists (Table 4.8).
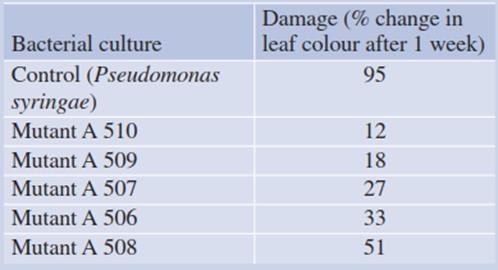
Table 4.8. Antagonists (mutants) of ice nucleation-active (INA) bacteria inhibiting noxious ice formation during moderate frost. (After Lindow (1982)). Pear leaves were inoculated with the respective bacterial culture 3 weeks prior to frost to give the bacteria time to establish themselves. Frost damage was quantified by assessing necrosis
These are obtained by plating out wash solutions from leaves and cooling them to —5 or —9 °C. Antagonists of INA bacteria should be present wherever ice formation does not take place. These organisms are mostly natural mutants of the original bacterial strains.
INA bacteria have found several technical and biotechnical applications: artificial snow making, cloud seeding and rain making in warmer regions, and use as part of a fusion protein of the N-terminus with a protein of interest (e.g. an enzyme (Gao et al. 2014)) or binding of cancer cells by an antibody or antibody mimetic protein displayed on the surface of engineered Escherichia coli cells (Zahnd et al. 2007).
Date added: 2025-01-17; views: 390;
