Defence Against Viruses via Gene Silencing
Defence against the systemic spread of viruses is conferred not only by the R gene-mediated hypersensitive response or by inducible systemic resistance; in fact, the main immune system against viruses in plants is based on gene silencing. For decades it has been observed in the field and under laboratory conditions that virus-infected plants develop resistance against a second infection by the same virus or a closely related virus. Weeks after an infection the newly developed leaves remain symptom-free even when inoculated with virus particles (Soosaar et al. 2005).
This phenomenon, called “recovery”, resembles the immunisation of humans or other mammals through treatment with an inactivated virus or a harmless related virus. We now know that a gene-silencing mechanism is behind the recovery from virus infection (Baulcombe 2004). This silencing targets virus- derived double-stranded RNA (dsRNA), which also explains why resistance covers not only the originally infecting virus but also viruses with a genome that is similar in sequence.
Most plant viruses are RNA viruses—that is, they carry genetic information as either single-stranded RNA (ssRNA) or double- stranded RNA instead of DNA. Different processes during the infection process give rise to the existence of dsRNA even when the genome is DNA or ssRNA. Replication catalysed by RNA-dependent RNA polymerases involves the formation of double-stranded intermediates. ssRNA can fold back and base pair into dsRNA. Bidirectional transcription of viral DNA can produce overlapping transcripts (Fig. 8.17).

Fig. 8.17. Virus-induced gene silencing. The presence of double-stranded RNA (dsRNA) during the replication of viral genomes triggers processing by RNase-like plant enzymes (Dicer and Dicer-like (DCL)). The resulting small RNAs are loaded into Argonaute (AGO)-containing RNA-induced silencing complexes (RISCs) to guide translational inhibition and/or slicing of viral RNA. The virus-derived small RNAs have the potential to spread through plasmodesmata to neighbouring cells, thereby activating a systemic antiviral defence (= immunisation). On green background the RNA silencing triggered by RNA viruses is depicted. Brown background and yellow symbols indicate the additional defence against DNA viruses via the modification of their DNA genomes (meth- ylation of DNA or histones) by small RNAs. Red symbols indicate viral suppressors of RNA silencing, produced by viruses to counteract the antiviral defence of the plant. P plasmodesmata. (Incarbone and Dunoyer 2013)
The presence of dsRNA triggers cleavage into small interfering RNAs (siRNAs) with a length of 21-24 nucleotides. The cleavage is catalysed by endonucleases called Dicer-like. Discovery of siRNAs in virus-infected plants was a major step in unravelling the existence of a complex world of regulatory small RNAs (Hamilton and Baulcombe 1999). One of the strands of the siRNA is then loaded onto another endonuclease, Argonaute (AGO), which is part of the so-called RNA-induced silencing complex (RISC). RISC targets RNA similar in sequence to the siRNAs and causes its degradation, thereby inhibiting replication or transcription of viral RNA. An alternative silencing mechanism is the sequence-specific methylation of DNA directed by siRNA. This mechanism is important for resistance against DNA viruses.
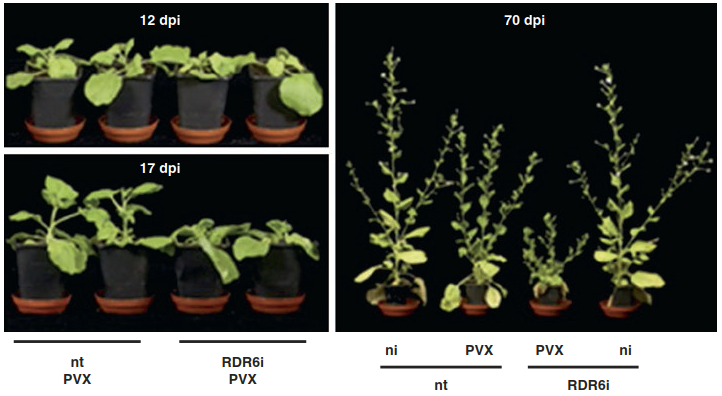
Fig. 8.18. Spreading of gene silencing confers resistance against viruses. Non-transformed Nicotiana benthaminana plants (nt) and plants with strongly reduced expression of the RNA-dependent RNA polymerase RDR6 (RDR6i) were inoculated with potato virus X (PVX). RDR6 is required for the response of cells to the systemic silencing signal. Stunting of RDR6i plants compared with nt and non-inoculated plants (ni) indicates loss of virus resistance. dpi days post infection
The protection of newly developed leaves requires the spreading of the RNA silencing throughout the plant. The action of RNA-dependent RNA polymerases on single-stranded viral RNA results in a strong amplification of the siRNAs. They can move through plasmodesmata and the phloem. In receiving cells they then trigger RNA-dependent RNA polymerases to produce siRNAs (Fig. 8.18). Viruses in turn have independently evolved a variety of proteins that suppress dsRNA-dependent virus resistance by interfering with components of the RNA silencing pathway. For example, suppressor protein p19, encoded in the genome of the tomato bushy stunt virus, binds to siRNA and prevents its integration into RISC. Thus, the corresponding viral RNAs can no longer be targeted for degradation (Fig. 8.17).
Date added: 2025-02-01; views: 359;
